Abstract
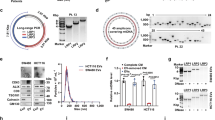
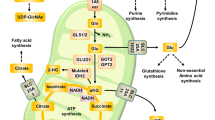
Cancer cells have metabolic features that allow them to preferentially metabolize glucose through aerobic glycolysis, providing them with a progression advantage. However, microRNA (miRNA) regulation of aerobic glycolysis in cancer cells has not been extensively investigated. We addressed this in the present study by examining the regulation of miR-139-5p on aerobic glycolysis of hepatocellular carcinoma (HCC) using clinical specimens, HCC cells, and a mouse xenograft model. We found that overexpressing miR-139-5p restrained aerobic glycolysis, suppressing proliferation, migration, and invasion in HCC cells. miR-139-5p regulated hexokinase 1 (HK1) and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) expression by directly targeting the transcription factor ETS1, which bound to the promoters of the HK1 and PFKFB3 genes. miR-139-5p-induced aerobic glycolysis, proliferation, migration, and invasion were reversed by ETS1 overexpression, while ETS1 silencing induced the expression of miR-139-5p via a post-transcriptional regulation mode involving Drosha. miR-139-5p expression was reduced in HCC compared to para-carcinoma tissue, which was confirmed in The Cancer Genome Atlas and GSE54751 HCC cohorts. Notably, the lower expression of mir-139 was correlated with worse prognosis. These outcomes indicate that reciprocal regulatory interactions between miR-139-5p and ETS1 modulate aerobic glycolysis, proliferation, and metastasis in HCC cells, suggesting new targets for HCC treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Endig J, Buitrago-Molina LE, Marhenke S, Reisinger F, Saborowski A, Schutt J, et al. Dual role of the adaptive immune system in liver injury and hepatocellular carcinoma development. Cancer Cell. 2016;30:308–23.
Tsoulfas G, Kawai T, Elias N, Ko SC, Agorastou P, Cosimi AB, et al. Long-term experience with liver transplantation for hepatocellular carcinoma. J Gastroenterol. 2011;46:249–56.
Zeng S, Zhang Y, Ma J, Deng G, Qu Y, Guo C, et al. BMP4 promotes metastasis of hepatocellular carcinoma by an induction of epithelial–mesenchymal transition via upregulating ID2. Cancer Lett. 2017;390:67–76.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Warburg O, On the origin of cancer cells. Sci (New York, NY). 1956;123:309–14.
Nie H, Li J, Yang XM, Cao QZ, Feng MX, Xue F, et al. Mineralocorticoid receptor suppresses cancer progression and the Warburg effect by modulating the miR-338-3p-PKLR axis in hepatocellular carcinoma. Hepatol (Baltim, Md). 2015;62:1145–59.
Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation. Sci (New York, NY). 2009;324:1029–33.
Hu J, Sun T, Wang H, Chen Z, Wang S, Yuan L, et al. MiR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell. 2016;29:49–60.
Nucera S, Giustacchini A, Boccalatte F, Calabria A, Fanciullo C, Plati T, et al. miRNA-126 orchestrates an oncogenic program in B cell precursor acute lymphoblastic leukemia. Cancer Cell. 2016;29:905–21.
Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Sci (New York, NY). 2010;328:1570–3.
Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22.
Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2016;30:349–51.
Liu L, Wang Y, Bai R, Yang K, Tian Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1alpha regulation. Oncogenesis. 2017;6:e318.
Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T, et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatol (Baltim, Md). 2015;62:1132–44.
Ma X, Li C, Sun L, Huang D, Li T, He X, et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat Commun. 2014;5:5212.
Li L, Kang L, Zhao W, Feng Y, Liu W, Wang T, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017;400:89–98.
Shen K, Mao R, Ma L, Li Y, Qiu Y, Cui D, et al. Post-transcriptional regulation of the tumor suppressor miR-139-5p and a network of miR-139-5p-mediated mRNA interactions in colorectal cancer. FEBS J. 2014;281:3609–24.
Rupaimoole R, Wu SY, Pradeep S, Ivan C, Pecot CV, Gharpure KM, et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun. 2014;5:5202.
Chabre O, Libe R, Assie G, Barreau O, Bertherat J, Bertagna X, et al. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr Relat Cancer. 2013;20:579–94.
Wong CC, Wong CM, Tung EK, Au SL, Lee JM, Poon RT, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–31.
Xu K, Shen K, Liang X, Li Y, Nagao N, Li J, et al. MiR-139-5p reverses CD44 + /CD133 + -associated multidrug resistance by downregulating NOTCH1 in colorectal carcinoma cells. Oncotarget. 2016;7:75118–29.
Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai S, et al. miR-139-5p inhibits the epithelial–mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep. 2016;6:27157.
Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–7.
Haakensen VD, Nygaard V, Greger L, Aure MR, Fromm B, Bukholm IR, et al. Subtype-specific micro-RNA expression signatures in breast cancer progression. Int J Cancer. 2016;139:1117–28.
Corbetta S, Vaira V, Guarnieri V, Scillitani A, Eller-Vainicher C, Ferrero S, et al. Differential expression of microRNAs in human parathyroid carcinomas compared with normal parathyroid tissue. Endocr Relat Cancer. 2010;17:135–46.
Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, Suzuki T, et al. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–9.
Emmrich S, Engeland F, El-Khatib M, Henke K, Obulkasim A, Schoning J, et al. miR-139-5p controls translation in myeloid leukemia through EIF4G2. Oncogene. 2016;35:1822–31.
Huang Y, Chen HC, Chiang CW, Yeh CT, Chen SJ, Chou CK. Identification of a two-layer regulatory network of proliferation-related microRNAs in hepatoma cells. Nucleic Acids Res. 2012;40:10478–93.
Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatol (Baltim, Md). 2012;56:622–31.
Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv Cancer Res. 2013;119:1–61.
He G, Tolic A, Bashkin JK. Heterogeneous dynamics in DNA site discrimination by the structurally homologous DNA-binding domains of ETS-family transcription factors. Nucleic Acids Res. 2015;43:4322–31.
Wang C, Kam RK, Shi W, Xia Y, Chen X, Cao Y, et al. The proto-oncogene transcription factor Ets1 regulates neural crest development through histone deacetylase 1 to mediate output of bone morphogenetic protein signaling. J Biol Chem. 2015;290:21925–38.
Dong Z. Acetylation of Ets-1 is the key to chromatin remodeling for miR-192 expression. Sci Signal. 2013;6:pe21.
Zhao J, Lu B, Xu H, Tong X, Wu G, Zhang X, et al. Thirty-kilodalton Tat-interacting protein suppresses tumor metastasis by inhibition of osteopontin transcription in human hepatocellular carcinoma. Hepatol (Baltim, Md). 2008;48:265–75.
Adam M, Schmidt D, Wardelmann E, Wernert N, Albers P. Angiogenetic protooncogene ets-1 induced neovascularization is involved in the metastatic process of testicular germ cell tumors. Eur Urol. 2003;44:329–36.
Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–62.
Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38.
Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34.
Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24.
Bao W, Fu HJ, Xie QS, Wang L, Zhang R, Guo ZY, et al. HER2 interacts with CD44 to up-regulate CXCR4 via epigenetic silencing of microRNA-139 in gastric cancer cells. Gastroenterology. 2011;141:2076. e2076.
Shen J, LeFave C, Sirosh I, Siegel AB, Tycko B, Santella RM. Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas. BMC Med Genom. 2015;8:28.
Acknowledgements
This work was supported by Nation’s Nature Science Foundation of China (Grant nos. 81472711, 91540111, 81672756, 81401180, and 81372283) and the Guangdong Province Nature Science Foundation (Grant no. 2014A030311013).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare they have no competing interests.
Rights and permissions
About this article
Cite this article
Hua, S., Lei, L., Deng, L. et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene 37, 1624–1636 (2018). https://doi.org/10.1038/s41388-017-0057-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-017-0057-3
This article is cited by
-
EZH2-H3K27me3-mediated silencing of mir-139-5p inhibits cellular senescence in hepatocellular carcinoma by activating TOP2A
Journal of Experimental & Clinical Cancer Research (2023)
-
GTF2E2 is a novel biomarker for recurrence after surgery and promotes progression of esophageal squamous cell carcinoma via miR-139-5p/GTF2E2/FUS axis
Oncogene (2022)
-
Ets1 mediates sorafenib resistance by regulating mitochondrial ROS pathway in hepatocellular carcinoma
Cell Death & Disease (2022)
-
MiR-652-5p elevated glycolysis level by targeting TIGAR in T-cell acute lymphoblastic leukemia
Cell Death & Disease (2022)
-
RNA splicing: a dual-edged sword for hepatocellular carcinoma
Medical Oncology (2022)