Abstract
Genetic analyses for bipolar disorder (BD) have achieved prominent success in Europeans in recent years, whereas its genetic basis in other populations remains relatively less understood. We herein report that the leading risk locus for BD in European genome-wide association studies (GWAS), the single-nucleotide polymorphism (SNP) rs9834970 near TRANK1 at 3p22 region, is also genome-wide significantly associated with BD in a meta-analysis of four independent East Asian samples including 5748 cases and 65,361 controls (p = 2.27 × 10−8, odds ratio = 1.136). Expression quantitative trait loci (eQTL) analyses and summary data-based Mendelian randomization (SMR) analyses in multiple human brain samples suggest that lower TRANK1 mRNA expression is a principal BD risk factor explaining its genetic risk signals at 3p22. We also identified another SNP rs4789 in the 3′ untranslated region (3′UTR) of TRANK1 showing stronger eQTL associations as well as genome-wide significant association with BD. Despite the relatively unclear neuronal function of TRANK1, our mRNA expression analyses in the human brains and in rat primary cortical neurons reveal that genes highly correlated with TRANK1 are significantly enriched in the biological processes related to dendritic spine, synaptic plasticity, axon guidance and circadian entrainment, and are also more likely to exhibit strong associations in psychiatric GWAS (e.g., the CACNA1C gene). Overall, our results support that TRANK1 is a potential BD risk gene. Further studies elucidating its roles in this illness are needed.
Similar content being viewed by others
Introduction
Bipolar disorder (BD) is a highly heritable neuropsychiatric disorder [1,2,3]. Genome-wide association studies (GWAS) in populations of European origins have identified numerous BD-associated genomic loci spanning CACNA1C, TRANK1, FADS1, MAD1L1, ANK3, NCAN, etc. [4,5,6,7,8,9,10]. Thanks to the currently available data of large BD case–control studies, we and others have confirmed single-nucleotide polymorphisms (SNPs) spanning FADS1, MAD1L1, and NCAN to be associated with BD in East Asians [11,12,13], suggesting the existence of shared genetic risk factors of BD between continental populations, and additional risk loci in East Asians are yet to be identified in larger samples.
Among the BD risk genes, TRANK1 has caught worldwide attention. An SNP rs9834970 downstream of TRANK1 showed genome-wide significant associations with BD in several GWAS of partially overlapped European cohorts [6,7,8, 14]. More recently, the Psychiatric Genomics Consortium (PGC2) performed a meta-analysis of multiple GWAS datasets including 20,352 BD patients and 31,358 controls from European populations, followed by replication analyses in an additional cohort of 9412 cases and 137,760 controls [4]. This PGC2 GWAS yielded 30 loci showing genome-wide associations with BD, and rs9834970 exhibited the strongest association signal [4]. While cumulative evidence suggests that rs9834970 is likely an authentic risk variation for BD, this contention is more persuasive in European populations. Whether this SNP confers genetic risk of BD in other ethnic populations and the potential mechanisms by which it participates in the illness remain undetermined.
In this study, we have replicated the association between rs9834970 and BD in 1784 patients and 2474 control subjects from Mainland China (p = 0.00218, odds ratio (OR) = 1.149 for C-allele). We then retrieved the statistical data from an independent Han Chinese BD sample [7, 15] and two Japanese BD samples [11] to perform a meta-analysis combining all these East Asian subjects (a total of 5748 cases and 65,361 controls) and observed a genome-wide significant association between rs9834970 and BD (p = 2.27 × 10−8, OR = 1.136 for C-allele). The rs9834970 risk C-allele indicated a lower level of TRANK1 mRNA in independent brain samples, which is in line with a recent study using human induced pluripotent stem cell (hiPSC)-derived neural cells [16]. Our results provide further evidence supporting TRANK1 as a lead risk gene for BD, and further studies elucidating the pathophysiological mechanisms mediated by this locus are therefore needed.
Materials and methods
The study protocol was approved by the ethics committee of the Second Affiliated Hospital of Xinxiang Medical University and the ethics committees of all participating hospitals and institutes. All participants provided written informed consents before any study-related procedures were performed.
Primary BD case–control sample
A total of 1784 BD cases and 2474 controls of Han Chinese origin were recruited from either mental health centers or psychiatric departments in general hospitals across cities of multiple provinces (i.e., Shanghai, Jiangsu, Zhejiang, Henan, and Hubei) in Mainland China. Part of the samples has been reported previously [12, 13, 17]. Each patient was independently interviewed and diagnosed as having BD by two research psychiatrists via an Extensive Clinical Interview and a Structured Clinical Interview for DSM-IV Axis/Disorders, Patient Version. Cases with pre-existing history of schizophrenia, mental retardation, and/or drug/alcohol addiction were excluded. Approximately 72% of the participating BD patients were affected by bipolar I, and 28% were bipolar II. Controls were local healthy volunteers with no current serious illnesses or disabilities or any personal or family history (including first-, second-, and third-degree relatives) of major psychiatric or neurological disorders (BD, depression, schizophrenia, attention deficit hyperactivity disorder, and mental retardation) based on self-report data. In the cases, 43% were males and 57% were females, whereas in the controls, 49% were males and 51% were females. The mean age was 37.9 ± 15.4 years for cases and 35.3 ± 12.6 years for controls at the time of sample collection. Individuals included in the current study have not enrolled in any previously published BD GWAS of Han Chinese origin [15]. Genomic DNA of the cases and controls were isolated from the peripheral blood using high-salt extraction method. The DNA samples were randomly distributed in 96-well plates and genotyped using SNaPShot following a previous study [17]. All assays were performed blind to diagnosis or genotype. The genotyping results of Sanger sequencing (2 SNPs in 50 randomly selected samples, performed using an ABI 3130 automatic sequencer, Applied Biosystems) and SNaPShot assays reached 100% concordance.
Statistical analysis and population stratification analysis
Hardy–Weinberg equilibrium test was performed for the cases and controls using Pearson χ2 test with one degree of freedom. Additive logistic regression analyses between SNPs and BD, covaring sex, and three principal components (PCs) were conducted to calculate p values, ORs, and 95% confidence intervals using PLINK v1.9 [18]. To avoid false positive and/or negative association signals caused by potential population stratification, we performed principal components analysis (PCA) for the genotype data of >300 random SNPs using the EIGENSTRAT software [19]. We also obtained data of these SNPs from distinct populations in 1000 Genomes Project and found that these SNPs were sufficient for differentiating samples by ethnicity (Fig. S1) [20].
Meta-analysis of independent BD datasets
To perform a meta-analysis of the available BD GWAS, we retrieved the statistics of candidate SNPs from two BD GWAS in East Asian populations [11, 15]. The East Asian BD GWAS were, respectively, conducted in an independent Han Chinese sample (1000 cases and 1000 controls) [7, 15] and two different Japanese samples (Ikeda 01: 1545 cases and 7408 controls; Ikeda 02: 1419 cases and 54,479 controls) [11]. Data of the European PGC2 GWAS was pooled with the above datasets for trans-ethnic meta-analyses [4]. The values of OR and standard error (SE) of each dataset were retrieved, and the classical inverse variance weighted methods were utilized under the fixed-effect model.
Expression quantitative trait loci (eQTL) analyses in public brain datasets
To identify whether the BD risk SNPs affected gene expression in the human brains, we utilized three public datasets containing genome-wide SNP genotype data and polyA+ RNA-seq data of human brains: (1) CommonMind (n = 467, dorsolateral prefrontal cortex (DLPFC)) [21], (2) Brain xQTL (n = 494, DLPFC) [22], and (3) Genotype-Tissue Expression project (GTEx, n = 205, brain cortex) [23].
We retrieved the gene-level eQTL results adjusted with surrogate variable (SV) analysis from 467 European individuals included in the CommonMind dataset (https://www.synapse.org/#!Synapse:syn2759792/wiki/69613) established by the CommonMind Consortium [21]. The RNA-seq data were adjusted for diagnosis, institution, sex, disease onset age, PMI, RNA integrity number (RIN), RIN2, clustered LIB, and 20 SVs. The eQTL was calculated according to the formula: adjusted gene expression ~ SNP dosage + ancestry vectors + diagnosis. Detailed information can be found in the original study [21].
The Brain xQTL Serve (http://mostafavilab.stat.ubc.ca/xQTLServe/) contains data obtained from the postmortem DLPFC tissues. The authors employed Spearman’s rank correlation to estimate the association between each SNP and mRNA levels of genes after removing known and hidden confounding factors in 494 individuals. Detailed information can be found in the original publication [22].
From the GTEx dataset (https://www.gtexportal.org/, 2017-06-05_v8), we retrieved the eQTL summary results of cortex tissues from 205 unaffected controls. For eQTL analysis, linear regression was conducted between SNPs and normalized expression matrices, covaring top five genotyping PCs, gender, genotyping platforms, and other covariates identified by the probabilistic estimation of expression residuals method [24]. Detailed information can be found in the original report [23].
Summary data-based Mendelian randomization (SMR) integrative analyses
Previous studies have described the SMR analysis as an effective approach in identifying susceptibility genes for complex diseases. SMR analysis integrates the summary-level statistics from trait GWAS and eQTL datasets to test whether effect sizes of certain SNPs on a given phenotype is attributed to gene expression [25]. The current study utilized the multi-SNP-based SMR, which considers multiple SNPs at a cis-eQTL locus to avoid the potential bias caused by analyzing only a single SNP [26, 27]. SNPs and genes in the major histocompatibility complex extended region (chr6:26M-34M, hg19) were not included in the SMR analyses due to their extreme complexity. In the present SMR analysis, the threshold of eQTL p value was set to be 1.00 × 10−5, genome-wide statistics of PGC2 BD GWAS was applied [4], and genes showing strong heterogeneity (PHEIDI < 0.01) in SMR were removed from further analyses.
Replication of eQTL associations in Chinese brain samples
Amygdala tissues of 65 non-psychiatric individuals were obtained from the Chinese Brain Bank Center. The tissue collection was conducted through the human body donation program organized and implemented by the Wuhan Red Cross Society. Permissions were obtained from the donors themselves or from their relatives for brain autopsy and use of the brain tissues in research. Further details regarding the donor, collection and storage procedures, and relevant ethnical approval have been described in a previous study [28]. RNA extraction, cDNA synthesis, and quantitative real-time PCR (RT-qPCR) were performed as previously described [12, 29,30,31]. The primers used for amplifying the housekeeping gene RPS13 were 5′-CCCCACTTGGTTGAAGTTGA-3′ (forward) and 5′-CTTGTGCAACACCATGTGAA-3′ (reverse) and primers for TRANK1 were 5′-TGATGAAAACAGAGACCCGCT-3′ (forward) and 5′-CTGCTTTCCACATTTCCGAGA-3′ (reverse). The relative gene expression levels were presented as the means of −ΔCt for statistical analysis against genotypic groups, and the p values were calculated using one-way analysis of variance covaring for sex, age, and RIN.
Correlation analysis in public human brain RNA-seq datasets
We downloaded the RNA-seq bam files (aligned to hg19) of non-psychiatric controls in BrainGVEX and CommonMind datasets from PsychENCODE website (https://www.synapse.org/#!Synapse:syn4921369) [21, 32]. The quantification of reads counts and gene expression were described in the previous study [27]. We excluded any genes with the average transcripts per million reads (TPM) < 2. For each gene, their TPM expression levels were log2 transformed and then underwent the Pearson’s analysis to estimate their correlation with TRANK1.
Trank1 short hairpin RNA (shRNA) constructs
The lentiviral vector pSicoR-Ef1a-mCh-Puro was used for knocking down the expression of Trank1 in rat neurons. Vectors containing two gene-specific shRNA sequences designed for rat Trank1 gene were constructed and verified through Sanger sequencing. The targeting sequences were Trank1-shRNA#1, 5′-GCGGCAGAATTGCTCAACA-3′; Trank1-shRNA#2, 5′-CACATTGATGCACCTCCTC-3′. The control shRNA sequence was: 5′-GATTTGCTGTTCGCCCAAG-3′.
Animals
Wild-type Sprague Dawley rats were purchased from Chengdu Dossy Experimental Animals CO., LTD (http://m.cd-dossy.cn/) and then bred in house. They were allowed free access to food and water on a 12 h light/dark cycle (lights on at 08:00 hours, lights off at 20:00 hours) with temperature (22 °C) and humidity (50–60%) controlled. All experiments were performed following the guidelines for ethical conduct in the care and use of animals (National Advisory Committee For Laboratory Animal Research).
Rat cortical neuronal cultures
Pregnant Sprague Dawley rats (E18/19) were euthanized using a CO2 chamber. Dissociated cortical neurons were prepared from the embryos following previous studies [33, 34]. In brief, cortices were dissected, trypsinized, and gently triturated into single-cell suspension. Neurons in this suspension were then counted and seeded at a density of 1 × 106 viable cells/well in 6-well culture plates previously coated with poly-D-lysine (10 μg ml−1) for at least 12 h at 37 °C. Cultures were maintained at 37 °C with 5% CO2, supplemented with Neurobasal medium with 2% B27 (Invitrogen) and 2.0 mM glutamax.
RNA-seq and differential expression analysis in rat neurons
Lentiviruses of pSicoR-Ef1a-mCh-Puro vector carrying Trank1-shRNA#1, Trank1-shRNA#2, or the empty vector were generated according to the manufacturer’s protocol. Rat neurons were infected at 14–15 days in vitro with Trank1-shRNAs or control shRNA. Neurons were harvested for RNA extraction and RNA-seq analysis 72 h post viral infection. Except for different reference genome used (GRCh38 for human was replaced by Rn6 for rat), the preprocessing, alignment, and quantification of RNA-seq data were conducted as previously described [29]. The raw RNA-Seq data in Trank1 knockdown rat neurons has been deposited in the Gene Expression Omnibus repository (accession number GSE153638). For differential expression analysis, DESeq2 [35] was used to analyze the expression differences between groups and to estimate size factors. Genes with the average fragments per kilobase million reads <1 in rat neurons were excluded from further analyses. Significantly differentially expressed genes (DEGs) were identified when false discovery rate (FDR)-corrected p value was <0.1.
Protein–protein interaction (PPI), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and Gene Ontology (GO) term analysis
Physical PPIs among genes of interest were constructed using the STRING database (version 11.0) [36] and CytoScape [37]. For pathway analysis, we performed functional annotation analyses (with KEGG pathway annotation and GO annotation) using R package clusterProfiler [38] to investigate whether the candidate genes were enriched in specific functional categories. Significantly enriched KEGG pathways and GO terms were identified when FDR-corrected p value was <0.05.
Results
Rs9834970 is genome-wide significantly associated with BD in East Asian populations
In our primary Han Chinese sample including 1784 BD cases and 2474 controls, rs9834970 was in Hardy–Weinberg equilibrium (Table S1), and its putative BD risk C-allele was overrepresented in patients compared with controls (p = 0.00218, OR = 1.149, Table S2). Through PCA, we confirmed that no obvious population stratification existed between our BD cases and controls (Fig. S1), suggesting that the observation was unlikely a false-positive signal caused by population stratification. We then retrieved the statistics of rs9834970 from an independent Han Chinese sample (1000 cases and 1000 controls) in a previous study [15] to validate our finding and found that the C-allele of rs9834970 was also associated with increased risk of BD (p = 0.0130, OR = 1.174, Table S2). It should be noted that there was no overlap between our primary BD case–control sample and this Chinese GWAS sample [15], as our primary Chinese sample was collected from Mainland China while subjects recruited for their study were not. The meta-analysis using two independent Han Chinese BD samples observed a stronger association (p = 8.15 ⨯ 10−5, OR = 1.158).
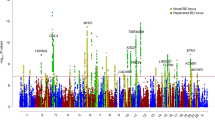
As mentioned earlier, a recent Japanese GWAS has also reported association between rs9834970 and BD in two independent samples (Ikeda 01: p = 9.41 × 10−4, OR = 1.150 in 1545 cases and 7408 controls; Ikeda 02, p = 0.0202, OR = 1.100 in 1419 cases and 54,479 controls; Table S2) [11]. The authors of these previous GWAS studies also demonstrated negligible population stratification in their samples [11, 15]. When all these East Asian samples (a total of 5748 cases and 65,361 controls) were pooled for a meta-analysis, rs9834970 was genome-wide significantly associated with BD (p = 2.27 × 10−8, OR = 1.136, Table S2). The sample size for our meta-analysis is comparable to other large-scale association studies in BD and psychiatric disorders [5,6,7,8,9], and it is reasonable to conclude that rs9834970 likely confers robust genetic risk of BD in East Asians. Given that rs9834970 has been initially discovered as a genome-wide significant marker for BD in populations of European ancestry (p = 5.72 × 10−12, OR = 1.080 in 29,764 cases and 169,118 controls) [4], we examined its linkage disequilibrium (LD) SNPs in Europeans and East Asians in the 1000 Genomes Project [20] and found that the LD patterns of this genomic area were similar between the two populations (Fig. S2). A subsequent meta-analysis of data from multiple ethnic populations was performed (including a total of 35,512 cases and 234,479 controls), and the association significance was further strengthened (p = 4.72 × 10−18, OR = 1.091, Table S2 and Fig. 1a).
Rs9834970 is associated with mRNA expression of TRANK1 and additional genes at 3p22 in public brain eQTL datasets
Accumulating studies suggest that GWAS risk loci of complex diseases tend to affect mRNA expression of nearby genes in relevant tissues and thereby contribute to the illnesses [39, 40]; we thus speculated that rs9834970 and its LD-linked SNPs might be associated with mRNA expression of neighboring genes at 3p22 in the brain. Through a quick exploration in the CommonMind DLPFC eQTL dataset (N = 467) [21], we found that rs9834970 was significantly associated with mRNA expression of TRANK1 (PeQTL = 2.84 × 10−6, Fig. 2a and Table S3), LRRFIP2 (PeQTL = 5.13 × 10−3), and DCLK3 (PeQTL = 1.88 × 10−5). We then examined this eQTL associations in independent brain eQTL datasets. In the Brain xQTL database of 494 European individuals [22], rs9834970 was also significantly associated with a lower mRNA level of TRANK1 (PeQTL = 0.00241, Fig. 2b and Table S4) rather than with other genes at 3p22. In the brain cortex tissues (N = 205) from GTEx dataset [23], rs9834970 was significantly associated with mRNA expression of TRANK1 (PeQTL = 5.90 × 10−3, Fig. 2c and Table S5) and DCLK3 (PeQTL = 5.60 × 10−3). These results suggest that rs9834970 is likely an eQTL SNP of surrounding genes at the 3p22 region.
a–c eQTL associations of rs9834970 with TRANK1 mRNA expression in CommonMind, Brain xQTL, and GTEx-cortex datasets. d–f eQTL associations of rs4789 with TRANK1 mRNA expression in CommonMind, Brain xQTL, and GTEx-cortex datasets. g, h eQTL associations of rs9834970 and rs4789 with TRANK1 mRNA expression in Han Chinese sample. CommonMind dataset, N = 467 individuals. Brain xQTL dataset, N = 494 individuals. GTEx-cortex dataset, N = 205 individuals. Han Chinese sample, N = 65 individuals.
SMR integrative analyses of brain eQTL datasets confirm TRANK1 as a BD susceptibility gene linked with genetic risk
To further depict the genetic risk architecture of the 3p22 region and to prioritize BD risk genes, we applied a multi-SNP-based SMR method to test the associations between risk of BD and mRNA expression of 3p22 region genes in the above three brain eQTL datasets (CommonMind, Brain xQTL, GTEx-cortex) [21,22,23]. According to the developer of this method, it can effectively prioritize susceptibility genes whose expression levels are relevant to the illness through assessing multiple SNPs at a cis-eQTL locus [26] and thereby define genes associated with BD genetic risk SNPs. In the CommonMind and Brain xQTL datasets, we found that TRANK1 was one of the top genes at the genome-wide level (PSMR-multi = 1.24 × 10−4 and 5.63 × 10−4, respectively, Fig. 3) and was the only BD risk gene at 3p22 (other 3p22 region genes were not significant). In the GTEx-cortex dataset, TRANK1 again showed significant association with risk of BD (PSMR-multi = 4.39 × 10−4, Fig. 3), while another nearby gene GOLGA4 was also significant (PSMR-multi = 3.72 × 10−3). Overall, the SMR integrative analysis suggested that TRANK1 mRNA expression likely contributed at least in part to the BD clinical risk associations at the 3p22 region.
Identification of an additional eQTL SNP rs4789 showing significant association with BD
During the SMR analysis, an additional SNP rs4789 in the 3′ untranslated region of TRANK1 was highlighted. Rs4789 is in substantial LD with rs9834970 in both European and East Asian populations (r2 ~ 0.8, Fig. S3) and was also genome-wide significantly associated with BD in PGC2 GWAS of European populations (p = 3.74 × 10−11, OR = 1.093 in 20,352 cases and 31,358 controls) [4]. We then examined its correlation with BD in Eastern Asian subjects. In our primary Han Chinese sample (1784 cases and 2474 controls), rs4789 was significantly associated with BD, with the major C-allele indicating an increased risk of the illness (p = 0.00896, OR = 1.123, Table S6). Given that accumulating studies have reported specific disease-prone haplotypes of a series of SNPs in BD [12, 41, 42], we then performed the rs9834970-rs4789 haplotype analysis in our Han Chinese sample (Table S7) and identified a risk haplotype (C-T, p = 0.00142, 6.9% in cases versus 4.9% in controls) and a protective haplotype (T-T, p = 7.09 × 10−6, 35.3% in cases versus 40.1% in controls) significantly associated with BD. Also, there was a rare haplotype (T-C) significantly associated with an increased risk of BD (T-C, p = 1.23 × 10−7, 2.7% in cases versus 1.1% in controls). Global tests also supported the associations of these haplotypes with BD (global p = 9.43 ⨯ 10−12, Pearson’s test).
In addition, rs4789 was also associated with BD in the Japanese sample (Ikeda 01: p = 4.68 × 10−3, OR = 1.120 in 1545 cases and 7408 controls; Ikeda 02, p = 0.0977, OR = 1.070 in 1419 cases and 54,479 controls; Table S6) [11]. Another Han Chinese dataset containing rs4789 [15] was not accessible to us, and this sample was thus not included in subsequent studies. Briefly, when all available samples of multiple ancestries (25,100 cases and 95,719 controls) were combined to conduct a meta-analysis, rs4789 was again genome-wide significantly associated with BD (p = 8.49 × 10−15, OR = 1.096, Fig. 1b and Table S6). Notably, the magnitude of significance of the association between rs4789 and BD was lower than that of the association between rs9834970 and BD, probably owing to the smaller sample size used for analysis of rs4789 (120,819 versus 269,991 subjects).
We found that rs4789 was also significantly associated with mRNA expression of TRANK1 in the CommonMind DLPFC eQTL dataset [21] (PeQTL = 1.34 × 10−9, Fig. 2d), Brain xQTL database (PeQTL = 2.17 × 10−6, Fig. 2e) [22], and GTEx-cortex dataset (PeQTL = 9.20 × 10−4, Fig. 2f) [23], and its eQTL associations with TRANK1 mRNA expression were stronger than rs9834970 in all these brain datasets. Rs4789 was also associated with mRNA expression of LRRFIP2 (PeQTL = 1.64 × 10−3 in CommonMind dataset, Table S3) and DCLK3 (PeQTL = 7.53×10−4 in CommonMind dataset, Table S3; PeQTL = 1.50 × 10−3 in GTEx-cortex dataset, Table S5), despite that the eQTL associations of these genes were less significant compared with TRANK1.
The BD risk alleles of both rs9834970 and rs4789 consistently predicted reduced expression of TRANK1 across the public brain eQTL datasets, which mainly contain data collected from Europeans. Considering that both SNPs were also associated with risk of BD in East Asians, we then examined whether they served as eQTLs of TRANK1 in this population. Given that DLPFC tissues were not available to us at the designing stage of the current study, we performed analyses in amygdala tissues as an indirect replication. Briefly, 65 amygdala tissues from non-psychiatric Han Chinese donors were collected, and RT-qPCR was performed to generate the eQTL data. Since the size of the Chinese sample was relatively small, individuals carrying homozygous T-alleles of rs9834970 were compared with those carrying at least one risk C-allele for a greater statistical power. Rs9834970 was marginally associated with TRANK1 mRNA expression in these subjects (PeQTL = 0.089, Fig. 2g), whereas rs4789 showed stronger eQTL association (PeQTL = 0.031, Fig. 2h), and individuals carrying risk C-alleles (both heterozygotes and homozygotes) of both SNPs had lower TRANK1 mRNA levels compared with those carrying the homozygous T-alleles. Despite the smaller size of the Chinese amygdala sample, the consistent direction of allelic effects across all the above analyses suggests a reliable link between genetic risk at 3p22 and TRANK1 expression.
Genes correlated with TRANK1 in the human brains are enriched in pathways of synapses, circadian entrainment, and calcium signaling and have higher proportions of exhibiting significant associations in psychiatric GWAS
We performed preliminary exploration of TRANK1’s function in the brain by conducting mRNA correlation analyses to reveal potential relevance of TRANK1 to other genes. We first analyzed the genes correlated with TRANK1 in two independent human brain RNA-seq datasets (BrainGVEX and CommonMind) [21, 32]. Control subjects from the two datasets were used so as to prevent confounding effects of medical treatment in cases. We calculated mRNA expression and examined whether any genes showed significant correlations with TRANK1 mRNA expression (defined as having |r| > 0.90). In the BrainGVEX dataset of 59 healthy controls, a total of 3163 genes showed strong positive correlations with TRANK1 mRNA expression (r > 0.90); in the CommonMind dataset of 50 normal subjects, 3261 genes were positively correlated with TRANK1 (r > 0.90). No genes showed significant negative correlations (r < −0.90) with TRANK1 in either brain dataset. The two brain datasets shared 1742 correlated genes (COGs) that were strongly associated with TRANK1 mRNA levels (Fig. 4a).
Proteins of these 1742 COGs formed dense PPI networks (PPI enrichment p value < 1.00 × 10−16, Fig. 4a). KEGG pathway analyses of these 1742 COGs revealed significant enrichment in the pathways of circadian entrainment, dopaminergic synapse, calcium signaling, axon guidance, oxytocin signaling, GABAergic synapse, glutamatergic synapse, long-term potentiation, and others (FDR < 0.05, Fig. 4a and Table S8). GO analysis revealed significant enrichment of TRANK1 COGs in terms “dendrite development”, “synapse organization”, “dendritic spine development”, and “regulation of synaptic plasticity” (Table S9). Many of these pathways and related biological processes have been found altered in BD subjects, such as dysregulations in circadian rhythm and synaptic function [43,44,45,46,47,48].
We then sought to further confirm the link between TRANK1 and BD via testing whether its COGs exhibited higher likelihood to be highlighted in BD GWAS. We first examined whether the aforementioned 1742 TRANK1 COGs were enriched in the risk intervals/genes highlighted by BD GWAS [4] using INRICH [49] and observed significant enrichment under multiple GWAS p value thresholds (e.g., enriched corrected p = 0.0024 at GWAS p value threshold < 5.00 × 10−6, Fig. 5a). Since TRANK1 has also been reported as a robust risk gene for schizophrenia [50], we then conducted enrichment analysis of the 1742 TRANK1 COGs with risk intervals/genes identified by schizophrenia GWAS [50] and again found a significant enrichment (e.g., INRICH enriched corrected p = 0.0020 at GWAS p value threshold < 5.00 × 10−6, Fig. 5a). In the additional enrichment analysis of the 1742 TRANK1 COGs in GWAS of major depressive disorder [51] and Alzheimer’s disease [52], no significant enrichment was observed for these illnesses (Fig. 5a), which is as expected since TRANK1 is not genetically associated with the risk of either major depressive disorder or Alzheimer’s disease. We also confirmed significant enrichment of the 1742 TRANK1 COGs among risk genes in the GWAS of BD and schizophrenia using another program MAGMA [53] (MAGMA enriched p = 5.54 × 10−7 for BD and p = 3.13 × 10−19 for schizophrenia, Fig. 5b). We then performed a detailed examination of genes shared between TRANK1 COGs and GWAS-identified risk markers of BD or schizophrenia. Eight genes (CACNA1C, DFNA5, SFMBT1, WHSC1L1, ZNF592, STK4, EFTUD1P1, UBE2Q2P1) were simultaneously highlighted in “TRANK1 COGs”, “GWAS of BD”, and “GWAS of schizophrenia” categories under the criteria that their MAGMA gene-level p values being <5.00 ⨯ 10−6 (Fig. 5c); ten additional genes (FAF1, BRE, TMEM178B, SF3B2, KLC2, C11orf80, WSCD2, TERF2, ILF3, HLF) were overlapped only between “TRANK1 COGs” and “GWAS of BD”. Intriguingly, the expression of TRANK1 was highly correlated with CACNA1C (BrainGVEX: r = 0.97, p = 5.28 × 10−37; CommonMind: r = 0.96, p = 6.16 × 10−28), one of the most significant genes in BD and schizophrenia.
a INRICH enrichment analysis of TRANK1 COGs and downregulated DEGs with psychiatric GWAS risk genes, and significant enrichment were observed for GWAS risk genes of BD and schizophrenia. b MAGMA enrichment analysis of TRANK1 COGs with psychiatric GWAS risk genes. c A detailed examination of overlapped genes among TRANK1 COGs, BD GWAS risk genes, and schizophrenia GWAS risk genes. BD bipolar disorder, SCZ schizophrenia, MDD major depressive disorder, AD Alzheimer’s disease, COG correlation gene.
Knockdown of Trank1 in cultured rat neurons confirms its impact on synapses and other processes related to BD as well as GWAS risk of psychiatric disorders
Given that the coding sequences of human TRANK1 and rat Trank1 are relatively conserved (≈84% in similarity), we then sought to further confirm the TRANK1-involved pathways revealed by human brain mRNA correlation analyses in rat neurons. Briefly, Trank1 shRNA infection of rat cortical neurons followed by RNA-seq were carried out. Lentiviral constructs of two rat Trank1 shRNAs and a negative control shRNA were established to infect the wild-type rat neurons for 72h (n = 3 per condition). Infection efficiency was estimated to be >80%, and the reduction of Trank1 mRNA was confirmed using RT-qPCR and RNA-seq (Fig. S4). The mean expression of Trank1 was reduced by 43.4% and 35.7%, respectively, in cells infected with Trank1-shRNA#1 and Trank1-shRNA#2 relative to those infected with control shRNA (both FDR < 1.00 × 10−19). Through RNA-seq analysis, we found that the expression levels of 2571 genes exhibited significant differences between neurons infected with Trank1-shRNA#1 and control shRNA (fold change (FC) > 1.2 at an FDR < 0.1). When Trank1 expression was knocked down using Trank1-shRNA#2, the expression levels of 4625 genes were significantly altered compared with controls (FC > 1.2 at an FDR < 0.1). Further analyses identified 1100 DEGs affected by both Trank1 shRNAs in the same direction, among which 594 DEGs were downregulated and 506 DEGs were upregulated (Fig. 4b).
Since our mRNA correlation analyses in the human brains only revealed genes positively correlated with TRANK1 expression, we focused on the 594 downregulated DEGs after Trank1 was knocked down. We found that proteins of these 594 DEGs also formed dense PPI networks (PPI enrichment p value < 1.00 × 10−16, Fig. 4b), and KEGG pathway analyses revealed significantly enriched signals in circadian entrainment, dopaminergic synapse, axon guidance, and oxytocin signaling pathway (FDR < 0.05, Fig. 4b and Table S10). Gene set enrichment analysis further indicated that these downregulated DEGs were strongly enriched in the GO terms “regulation of synaptic plasticity”, “forebrain development”, “synaptic membrane”, “postsynaptic membrane”, “regulation of Wnt signaling pathway”, etc. (FDR < 0.05, Table S11). Collectively, our correlation analyses and shRNA knockdown experiments provided consistent results highlighting the involvement of TRANK1 in dendritic spines and synapses.
We further examined the enrichment of the 594 downregulated DEGs among risk intervals/genes identified by GWAS of BD and schizophrenia [4, 50] and also found a significant enrichment signal at a particular GWAS threshold (e.g., INRICH enriched corrected p = 0.038 for BD at GWAS p value threshold < 5.00 × 10−7, INRICH enriched corrected p = 0.0012 for schizophrenia at GWAS p value threshold < 5.00 × 10−6, Fig. 5a). Collectively, these results suggest that the function of TRANK1 may be related to other psychiatric risk genes.
Discussions
BD has been a severe threat to public well-being, but our knowledge regarding this illness has been limited, let alone effective prevention and management. Accumulating studies suggest that rs9834970 near TRANK1 is one of the best replicated risk SNPs for BD [4, 6,7,8, 11, 14]. In the current study, using an independent Han Chinese sample, we present further evidence for the association between rs9834970 and BD, and meta-analyses in four cohorts of East Asian individuals indicate that rs9834970 is indeed a BD risk SNP. We also identified another SNP rs4789 in considerable LD with rs9834970 showing genome-wide significant association with BD. This SNP has been mentioned in previous GWAS [6, 11], and the level of statistical significance was lower than that of rs9834970, which is also in line with our results in the Han Chinese samples. However, the lower significance level does not deny its potential impact on TRANK1 expression, which is reflected by its significant eQTL associations in independent brain samples. The haplotype analysis of rs9834970 and rs4789 further confirms significant association of TRANK1 with BD and suggests that this region may harbor additional causal risk variations or be affected by epistasis. Further fine-scale mapping analysis may answer this question. Notably, a recent study also identified another functional SNP rs906482 affecting TRANK1 expression and CTCF-binding affinities [16]. Overall, these results suggest that there may be more than one functional SNP in the TRANK1 locus.
Our eQTL results are consistent with a recent study [16], in which the risk allele of rs9834970 was significantly associated with lower expression of TRANK1 in hiPSC-derived neural progenitor cells. They also showed that decreased expression of TRANK1 perturbed the expression of many genes involved in neural development and differentiation, and the lower TRANK1 expression correlated with the rs9834970 BD risk allele could be rescued by chronic treatment with therapeutic dosages of valproic acid [16]. Therefore, lower expression of TRANK1 might be linked with genetic risk of BD. We have also shown that the association of rs4789 with lower TRANK1 expression is more significant in multiple brain samples, further confirming that lower TRANK1 expression is likely involved in BD pathogenesis.
In summary, we have presented convergent evidence for the association of TRANK1 with BD and have provided hints for the underlying biological processes. Further in vivo disruption of this gene followed by thorough pathophysiological analyses and examination of BD-relevant cognitive, emotional, and behavioral impairments is necessary for a better understanding of BD pathogenesis. While the involvement of genetic variations at 3p22 in BD is supported by the genetic analyses in the current study, lacking of large-scale GWAS in East Asians remains a major difficulty in the identification of additional BD risk loci. To gain a more comprehensive understanding of BD genetic architecture in this population, GWAS and fine-mapping analysis are necessary. In addition, the controls of our sample were recruited based on their self-report of no personal or family history of mental illnesses rather than psychiatric screening by professional staff, and a more stringent inclusion criteria for controls would be ideal. Finally, despite significant associations between genetic variations at 3p22 (e.g., rs9834970 and rs4789) and TRANK1 mRNA expression in multiple brain eQTL samples (e.g., CommonMind, Brain xQTL, GTEx-cortex, and our Chinese sample) and in hiPSC-derived neural cells [16], inconsistency still exists in some other datasets in which these SNPs are not associated with TRANK1 (such as BrainSeq [54] and PsychENCODE [32]), probably due to the differences in the sequencing platforms, analytical methods, and sample compositions (e.g., BrainSeq included many samples of African American ancestry) between different datasets. Nevertheless, it should be acknowledged that TRANK1 is unlikely the only gene explaining the association between BD clinical risk and the 3p22 locus. Identifying the involvement and pathological contribution of additional risk genes at 3p22 is necessary to fully elucidate the psychiatric risk components underlying this genomic region.
Funding and disclosure
The authors declare no competing financial interests. This work was supported by grants from National Natural Science Foundation of China (81871067 to H.C., 81671330 and 81971252 to L.L., U1904130 to W.L., 81771439 to L.H.); Open Program of Henan Key Laboratory of Biological Psychiatry (ZDSYS2018001 to H.C.); the Science and Technology Project of Henan Province (192102310086 to W.L.); the High Scientific and Technological Research Fund of Xinxiang Medical University (2017ZDCG-04 to L.L.); Hubei Province health and family planning scientific research project (WJ2015Q033 to N.Q.); Population and Family Planning Commission of Wuhan (WX14B34 to N.Q.); and the medical and health science and technology project in Zhejiang (2018KY721 to D.-S.Z.). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 11/16/2019. Data were generated as part of the PsychENCODE Consortium, supported by: U01MH103392, U01MH103365, U01MH103346, U01MH103340, U01MH103339, R21MH109956, R21MH105881, R21MH105853, R21MH103877, R21MH102791, R01MH111721, R01MH110928, R01MH110927, R01MH110926, R01MH110921, R01MH110920, R01MH110905, R01MH109715, R01MH109677, R01MH105898, R01MH105898, R01MH094714, P50MH106934, U01MH116488, U01MH116487, U01MH116492, U01MH116489, U01MH116438, U01MH116441, U01MH116442, R01MH114911, R01MH114899, R01MH114901, R01MH117293, R01MH117291, R01MH117292. The awardees of members in PsychENCODE Consortium are listed in Supplementary Materials. Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd, and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138, and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, and the NIMH Human Brain Collection Core. The members of CommonMind Consortium leadership are listed in Supplementary Materials.
References
Vieta E, Berk M, Schulze TG, Carvalho AF, Suppes T, Calabrese JR, et al. Bipolar disorders. Nat Rev Dis Prim. 2018;4:18008.
Gordovez FJA, McMahon FJ. The genetics of bipolar disorder. Mol Psychiatry. 2020;25:544–59.
Zhang C, Xiao X, Li T, Li M. Translational genomics and beyond in bipolar disorder. Mol Psychiatry. 2020. https://doi.org/10.1038/s41380-41020-40782-41389.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Cichon S, Muhleisen TW, Degenhardt FA, Mattheisen M, Miro X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–81.
Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landen M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25:3383–94.
Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2013;18:195–205.
Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5:3339.
Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8.
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83.
Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018;23:639–47.
Zhao L, Chang H, Zhou DS, Cai J, Fan W, Tang W, et al. Replicated associations of FADS1, MAD1L1, and a rare variant at 10q26.13 with bipolar disorder in Chinese population. Transl Psychiatry. 2018;8:270.
Wang L, Liu W, Li X, Xiao X, Li L, Liu F, et al. Further evidence of an association between NCAN rs1064395 and bipolar disorder. Mol Neuropsychiatry. 2018;4:30–4.
Goes FS, Hamshere ML, Seifuddin F, Pirooznia M, Belmonte-Mahon P, Breuer R, et al. Genome-wide association of mood-incongruent psychotic bipolar disorder. Transl Psychiatry. 2012;2:e180.
Lee MT, Chen CH, Lee CS, Chen CC, Chong MY, Ouyang WC, et al. Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol Psychiatry. 2011;16:548–56.
Jiang X, Detera-Wadleigh SD, Akula N, Mallon BS, Hou L, Xiao T, et al. Sodium valproate rescues expression of TRANK1 in iPSC-derived neural cells that carry a genetic variant associated with serious mental illness. Mol Psychiatry. 2019;24:613–24.
Li W, Yang Y, Luo B, Zhang Y, Song X, Li M, et al. Association of SYNE1 locus with bipolar disorder in Chinese population. Hereditas. 2019;156:19.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci. 2017;20:1418–26.
GTEx Consortium, Laboratory Data Analysis, Coordinating Center Analysis Working Group, Statistical Methods groups-Analysis Working Group, Enhancing GTEx groups, NIH Common Fund, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13.
Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:e1000770.
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7.
Wu Y, Zeng J, Zhang F, Zhu Z, Qi T, Zheng Z, et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun. 2018;9:918.
Li HJ, Qu N, Hui L, Cai X, Zhang CY, Zhong BL, et al. Further confirmation of netrin 1 receptor (DCC) as a depression risk gene via integrations of multi-omics data. Transl Psychiatry. 2020;10:98.
Wang Z, Wang N, Li Z, Xiao F, Dai J. Human high intelligence is involved in spectral redshift of biophotonic activities in the brain. Proc Natl Acad Sci USA. 2016;113:8753–8.
Li H, Chang H, Song X, Liu W, Li L, Wang L, et al. Integrative analyses of major histocompatibility complex loci in the genome-wide association studies of major depressive disorder. Neuropsychopharmacology. 2019;44:1552–61.
Liu W, Yan H, Zhou D, Cai X, Zhang Y, Li S, et al. The depression GWAS risk allele predicts smaller cerebellar gray matter volume and reduced SIRT1 mRNA expression in Chinese population. Transl Psychiatry. 2019;9:333.
Cai X, Yang ZH, Li HJ, Xiao X, Li M, Chang H. A human-specific schizophrenia risk tandem repeat affects alternative splicing of a human-unique isoform AS3MTd2d3 and mushroom dendritic spine density. Schizophr Bull. 2020. https://doi.org/10.1093/schbul/sbaa1098.
PsychEncode Consortium, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–12.
Li H, Zhou DS, Chang H, Wang L, Liu W, Dai SX, et al. Interactome analyses implicated CAMK2A in the genetic predisposition and pharmacological mechanism of bipolar disorder. J Psychiatr Res. 2019;115:165–75.
Yang Z, Zhou D, Li H, Cai X, Liu W, Wang L, et al. The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function and mushroom dendritic spine. Mol Psychiatry. 2020;25:48–66.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–97.
Gallagher MD, Chen-Plotkin AS. The post-GWAS era: from association to function. Am J Hum Genet. 2018;102:717–30.
Gonzalez SD, Xu C, Ramirez ME, Zavala JM, Armas R, Contreras SA, et al. Family-based association of an ANK3 haplotype with bipolar disorder in Latino populations. Transl Psychiatry. 2013;3:e265.
Liu J, Li ZQ, Li JY, Li T, Wang T, Li Y, et al. Polymorphisms and haplotypes in the YWHAE gene increase susceptibility to bipolar disorder in Chinese Han population. J Clin Psychiatry. 2012;73:e1276–82.
Nurnberger JI Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry. 2014;71:657–64.
Akula N, Wendland JR, Choi KH, McMahon FJ. An integrative genomic study implicates the postsynaptic density in the pathogenesis of bipolar disorder. Neuropsychopharmacology. 2016;41:886–95.
Konopaske GT, Lange N, Coyle JT, Benes FM. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 2014;71:1323–31.
Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–93.
Forrest MP, Parnell E, Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat Rev Neurosci. 2018;19:215–34.
Focking M, Dicker P, Lopez LM, Hryniewiecka M, Wynne K, English JA, et al. Proteomic analysis of the postsynaptic density implicates synaptic function and energy pathways in bipolar disorder. Transl Psychiatry. 2016;6:e959.
Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9.
Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21:1117–25.
Acknowledgements
We acknowledge with appreciation all the individuals with bipolar disorders and healthy controls whose contributions made this work possible. We are deeply grateful to all the participants working on this project. The authors wish to acknowledge Dr. Bernard Ng (Departments of Statistics and Medical Genetics, University of British Columbia, Vancouver, BC, Canada), Dr. Sara Mostafavi (Departments of Statistics and Medical Genetics, University of British Columbia, Vancouver, BC, Canada), and Dr. Philip L De Jager (Broad Institute, Cambridge, MA, USA) for providing the individual-level TRANK1 expression and genotypes in the Brain xQTL dataset.
Author information
Authors and Affiliations
Contributions
H.C., L.L., and M.L. conceived the study and interpreted the results. W.L., X.C., M.S., H.-J.L., L.Z., W.L., and L.W. performed the DNA extraction, SNP genotyping, statistical analysis, and the primary experiments. H.-J.L. and C.-Y.Z. performed the RNA-seq and pathway analysis. W.L., M.S., Y.Y., L.Z., M.S., Y.Z., C.Z., D.-S.Z., X.L., L.H., Q.-F.J., N.Q., B.-L.Z., S.-F.Z., J.C., B.X., Y.L., X.S., W.F., W.T., W.T., J.T., X.C., Y.F., and L.L. carried out subject recruitment and phenotype analysis. W.Y. and D.Z. contributed to design of sample analysis. X.X., H.C., L.L., and M.L. drafted the manuscript, and all authors contributed to the final version of the paper.
Corresponding authors
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, W., Cai, X., Li, HJ. et al. Independent replications and integrative analyses confirm TRANK1 as a susceptibility gene for bipolar disorder. Neuropsychopharmacol. 46, 1103–1112 (2021). https://doi.org/10.1038/s41386-020-00788-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41386-020-00788-4
This article is cited by
-
TrkB-dependent regulation of molecular signaling across septal cell types
Translational Psychiatry (2024)
-
Sleep Disorder Kleine–Levin Syndrome (KLS) Joins the List of Polygenic Brain Disorders Associated with Obstetric Complications
Cellular and Molecular Neurobiology (2023)
-
Multi-ancestry meta-analysis and fine-mapping in Alzheimer’s disease
Molecular Psychiatry (2023)
-
Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders
Nature Neuroscience (2022)
-
Phenotypes, mechanisms and therapeutics: insights from bipolar disorder GWAS findings
Molecular Psychiatry (2022)