Abstract
Investigations in recent decades have exploited tissue DNA genotyping as a powerful ancillary tool for the precision diagnosis and subclassification of gestational trophoblastic disease. As lesions of gestational origin, the inherited paternal genome, with or without copy number alterations, is the fundamental molecular basis for the diagnostic applications of DNA genotyping. Genotyping is now considered the gold standard in the confirmation and subtyping of sporadic hydatidiform moles. Although a precise diagnosis of partial mole requires DNA genotyping, prognostic stratification according to distinct genetic zygosity in complete moles has recently gained significant clinical relevance for patient care. Beyond hydatidiform moles, DNA genotyping has fundamental applications in the diagnosis or prognostic assessment of gestational trophoblastic tumors, in particular gestational choriocarcinoma. DNA genotyping provides a decisive tool in the separation of gestational trophoblastic neoplasia from non-gestational counterparts/mimics of either germ cell or somatic origin. The FIGO/WHO prognostic scoring scheme requires ascertaining the precise index gestational event and the time interval between the tumor and index gestation, where DNA genotyping can provide highly relevant information. With rapid acquisition of molecular diagnostic capabilities in the clinical practice, DNA genotyping has become closely integrated into the routine diagnostic workup of various forms of gestational trophoblastic disease.
Similar content being viewed by others
Introduction
In the advent of early clinical detection and intervention, gestational trophoblastic disease (GTD), notably hydatidiform moles, are evacuated at much earlier stages of gestation than in previous decades. Such early intervention has drastically changed the overall landscape of clinical presentation and pathological diagnosis of various subtypes of GTD [1, 2]. For hydatidiform moles, the classical clinical symptomatology—vaginal bleeding during second trimester and excessive uterine size—is uncommon and most patients now present with missed abortion during the first trimester. Subsequently, the characteristic morphologic features of molar gestations, both complete and partial moles (Figs. 1 and 2), are less well developed and overlap with many non-molar conditions. An estimated 50% of true partial moles cannot be accurately diagnosed by routine histomorphology and significant inter- and intra-observer variability exists even among expert pathologists [3, 4]. However, distinction of hydatidiform moles from non-molar specimens and the subclassification of hydatidiform moles are critical for risk assessment for the development of post-molar gestational trophoblastic neoplasia (GTN), which varies among subtypes of hydatidiform moles and so does the duration of required clinical follow-up. Complete moles progress into persistent/invasive mole in ~15–20% and into gestational choriocarcinoma in 2–3% of cases [5,6,7,8], whereas the risk after partial moles is 0.5–5% for persistent/invasive mole [9, 10] and 0.015% for choriocarcinoma [11]. It must be further emphasized that the current diagnostic inaccuracy, in particular of partial hydatidiform mole (PHM), continues to compromise scientific investigations into the epidemiology, pathogenesis, and biological behavior of hydatidiform moles due to inaccurately classified study cohorts.
Frequent findings of PHM include hydropic villi with cistern formation (upper left), abnormal villous shapes and contours (upper right), and trophoblastic pseudo-inclusions (lower left). P57 immunostain shows normal retained nuclear staining in cytotrophoblast and villous stromal cells (lower right).
There have been remarkable advances in our understanding of the genetic basis of GTD over the past decades, followed by more recent development of ancillary tools for accurate diagnosis and classification of GTD. DNA genotyping using PCR to analyze short-tandem repeat (STR) polymorphism has emerged as a powerful tool in the precise diagnosis and classification of hydatidiform moles. Over the past decade, the clinical sensitivity and specificity of DNA genotyping for the diagnosis and subclassification of hydatidiform moles has been confirmed by numerous studies [12,13,14,15,16]. Beyond hydatidiform moles, DNA genotyping has fundamental applications in the assessment of GTN. The presence of distinct paternal genetic profile by DNA genotyping effectively separates a gestational trophoblastic tumor from its non-gestational counterpart of either germ cell or somatic origin. DNA genotyping comparison between the gestational trophoblastic tumor and tissue samples of the prior pregnancy/pregnancies can determine the type of index gestation (term pregnancy, molar gestation, or non-molar abortion) and the time interval between the development of the tumor and its index gestation for the International Federation of Gynecology and Obstetrics/World Health Organization (FIGO/WHO) risk scoring scheme for clinical management of patients with GTN.
Genetic basis and principles of STR genotyping diagnosis of GTD
Normal gestational tissue inherits one haploid genomic complement from each of the parents, referred to as maternal and paternal haploid genome, respectively. Depending on the nature of gestational trophoblastic disease, the copy number of parental haploid complements varies. Determination of the copy number of the parental haploid genomes is the fundamental molecular basis for genotyping diagnosis of hydatidiform moles. Cytogenetic investigations in the 1970s were pivotal to elucidation of the pathogenesis of hydatidiform moles [17, 18]. In sporadic complete hydatidiform moles (CHMs), the gestational tissues inherit an androgenic-only nuclear genome and a maternal-only mitochondrial DNA [19, 20], with either 46XX diploid karyotypes (homozygous, 80–90%) or 46XX or XY diploid karyotypes (heterozygous, 10–20%) [18, 21]. Familial biparental CHM (FBCHM) is an exceptionally rare condition in sisters of affected families [22, 23], who suffer from recurrent complete moles. Inherited NLRP7/NALP7 (nucleotide-binding, leucine-rich repeat, Pyrin domains) [24] mutations of either homozygous or compound heterozygous manner are responsible for the majority of FBCHM [23,24,25,26]. NLRP7 is the first identified human maternal effect gene, genes that are expressed in the oocyte to support embryonic development until activation of the embryonic genome occurs [24]. Around 50 missense or truncating mutations of NLRP7 have been documented [27, 28]. The expression of the NRLP7 gene is autosomal recessive and is transcribed in the unfertilized oocyte at or before meiosis I. KHDC3L (KH domain containing 3-like) is the second maternal effect gene identified in FBCHM with four homozygous or compound heterozygous mutations responsible for the remaining 10–14% of cases [29, 30]. PHMs are triploid with a diandric-monogynic genome, arising from two paternal haploid sperms combined with an ovum (dispermic/heterozygous PHM) in >95% of cases and the remaining <5% of cases involve an ovum fertilized by either one haploid sperm followed by paternal haploid reduplication or one diploid sperm due to the failure of meiosis I or II (monospermic, homozygous PHM) [31, 32]. Consequently, an estimated 70% of PHMs have a 69XXY karyotype, 27% are 69XXX and 3% 69XYY [12, 32, 33]. Rare tetraploid PHMs have also been reported [34,35,36].
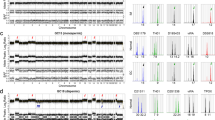
Genotyping provides a measurement of the genetic variation between members of a species. Various molecular targets have been explored in the past for human genotyping or identity testing (DNA restriction fragment length polymorphism, enzyme polymorphism, single-nucleotide polymorphism, and STR polymorphism). STRs are repetitive DNA sequences of two to seven nucleotides that are highly prevalent in the noncoding regions of the human genome and are genetically stable [37]. STR polymorphism denotes that alleles at the STR locus differ in the number of repeats between individuals and, therefore, the genetic identity can be distinguished from one another, usually by analyzing multiple STR loci [37,38,39]. By the same principle, STR genotyping of gestational tissue in comparison with corresponding maternal tissue can determine the parental genomic contribution in a hydatidiform mole. There are a number of robust, widely commercialized STR genotyping kits targeting at various multiple STR loci, such as AmpFlSTR® Identifiler™ (Applied Biosystem, Inc.) and PowerPlex® (Promega, Inc.). These assays resemble a conventional diagnostic PCR procedure, involving manual tissue dissection, DNA extraction, one multiplex PCR reaction, and capillary electrophoresis. As abnormal products of conceptions (POCs) are nowadays evacuated during the first trimester, the specimens likely do not have grossly identifiable villi. Genotyping is usually performed using formalin-fixed paraffin-embedded tissue samples. In general, DNA is extracted from manually scaped pure chorionic villi from unstained sections, guided by the corresponding hematoxylin and eosin-stained section. In rare situations where pure target tissue cannot be obtained by manual dissection, laser microdissection may be used. DNA of pure maternal decidua from each case is also extracted and genotyped for comparative analysis. In POCs without maternal decidua, the patient’s previously archived tissue samples or peripheral blood, or buccal swab samples may be used for comparative genotyping. Using as little as 1.5–2.0 ng template DNA (150–200 human diploid/chorionic villous cells), these STR assays produce PCR amplicons ranging from 100 to 350 base pairs, particularly suitable for DNA template extracted from formalin-fixed paraffin-embedded tissues [39, 40]. Over the past decade, the clinical sensitivity and specificity of STR genotyping analysis of POCs were confirmed by numerous studies [12,13,14,15,16, 41, 42]. Currently, STR genotyping has been established as the gold standard for the precise diagnosis and subclassification of sporadic hydatidiform moles (Fig. 3) [4]. For detailed glossary of terms used in this article see supplementary information (Supplementary information).
Each panel consists of comparative STR allelic patterns between gestational endometrium (Endo) and chorionic villous tissue (Villi). Complete mole demonstrates either monospermic/homozygous allelic pattern in the villi (CHM-MS, single homozygous allele at each STR locus) or dispermic/heterozygous allelic pattern in the villi (CHM-DS, asterisk indicating two distinct paternal alleles). Partial mole shows dispermic/heterozygous triploid allelic pattern (PHM-DS) or monospermic/homozygous triploid allelic pattern (PHM-MS, asterisk indicating two copies of the same paternal allele). Non-molar digynic–monoandric triploid gestation demonstrates two maternal alleles in addition to one paternal allele at all STR loci (Digynic Triploid). Trisomy 21 gestation shows the presence of three allelic copies at one STR locus on chromosome 21 (asterisk). Paternal uniparental disomy of chromosome 11 demonstrates two distinct paternal alleles at the THO1 locus on chromosome 11 (UPD11, asterisk indicating the THO1 locus). Normal gestation shows a balanced biparental genetic profile at all STR loci in the villi (Normal Gestation).
DNA genotyping is required for definitive diagnosis of PHM
PHMs manifest as a spectrum of morphological changes (Fig. 2) significantly overlapping with various non-molar gestations including those with chromosomal alterations, in particular chromosomal trisomies, digynic triploid conceptions, placental mesenchymal dysplasia, and hydropic abortions [43,44,45,46,47]. Gestations with trisomy (in particular trisomy 7, 8, 13, 15, 16, 18, 21, and 22) may simulate PHM by their abnormal villous configurations, trophoblastic pseudo-inclusions, and villous hydrops [45, 48, 49]. Trophoblastic hyperplasia may also be prominent, especially in trisomies involving chromosomes 7, 15, 21, and 22 (Fig. 4). Triploidy is one of the most common chromosomal abnormalities in human conceptions, occurring in up to 3% of all gestations [50, 51] and 8–10% of spontaneous abortions [52, 53]. An estimated one-third of triploid gestations are digynic non-molar, arising from meiotic non-disjunction of maternal chromosomes [32, 50, 54]. Although the proportion of digynic triploids undergoing comprehensive diagnostic workup is low (~3%) [12] due to the absence of clinical and histological features of hydatidiform moles, in rare cases, digynic triploidy can present with villous hydrops and, less commonly, cistern formation, irregular villous shape, and trophoblastic pseudo-inclusions (Fig. 4). Placental mesenchymal dysplasia evacuated during the second or third trimester may mimic PHM with villous hydrops, aneurysmal/hemangiomatous stem villous vessels. The villous stroma consists of spindle and stellate mesenchymal cells in a myxoid background. The fetus may show growth restriction or signs of Beckwith–Wiedemann syndrome [54]. Hydropic abortion usually demonstrates mild hydropic change but the villi have a smooth contour without circumferential trophoblastic hyperplasia or pseudo-inclusions (Fig. 4) [45]. Morphologic spectra of CHM and PHM can also overlap with regard to variations in villous size and shape (Fig. 5), the extent of hydropic change, and trophoblastic hyperplasia. Early gestations, in particularly at an ectopic site, may also show histological findings overlapping with PHM or CHM (Fig. 5). The distinction between partial and very early complete hydatidiform moles may be facilitated by the identification of fetal tissues including nucleated red blood cells, although the latter may rarely also be observed in very early CHM [55, 56].
Ancillary studies are essential for the diagnosis of PHM. Although flow cytometric DNA ploidy analysis has been traditionally used to facilitate the recognition by demonstrating triploidy [4, 43, 57, 58], the mere presence of triploidy is not diagnostic of PHM as a third of triploid cases are digynic–monoandric non-molar gestations [54]. In addition, ploidy analysis using paraffin-embedded tissue frequently suffers from technical difficulties and interpretation errors, resulting in a significant misclassification of ploidy [59]. As the morphologic features of PHM are shared frequently by numerous histological mimics, appropriate triaging of specimens for DNA genotyping is important. According to a recent comprehensive assessment of histology in correlation with DNA genotyping, all traditional morphologic parameters attributed to PHMs are nonspecific and are shared in a comparable proportion by various non-molar conditions including trisomic gestations and hydropic abortions [48]. The presence of at least one of the following three features—cistern formation (single cavity occupying at least 50% of the villous volume), two villous populations, round to oval trophoblastic pseudo-inclusions—coupled with terminal villous size ≥ 2.5 mm has a specificity of 84%, sensitivity of 61%, and positive predictive value (PPV) of 72% for PHM. The presence of both cistern formation and villous size ≥ 2.5 mm has the highest PPV of 90% with specificity of 96% [48], although the sensitivity is only 28%. As no single or combined morphological features are sufficient for the diagnosis of PHM, it is recommended that the presence of any one of the following microscopic findings should prompt DNA genotyping to rule out PHM: round or oval trophoblastic pseudo-inclusions, cistern formation, two populations of villi, and villous size of 2.5 mm or larger. It should be pointed out that if prenatal fetal testing indicates the presence of various trisomies, genotyping of POCs to rule out molar gestation may not be necessary. However, if the prenatal testing indicates fetal triploidy, genotyping is often required to rule out a partial mole. Given the precise analytical power of interrogating the parental genetic composition, DNA genotyping is now required for definitive diagnosis of partial mole according to the current WHO tumor classification [47].
Genotypical subclassification of complete moles is clinically relevant for patient management
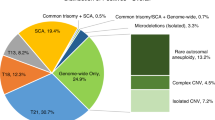
Although it has been well established that complete moles carry a significantly higher risk than partial moles for the development of post-molar gestational trophoblastic disease, the correlation between the genetic subtypes of complete moles and post-molar gestational trophoblastic disease has been a subject of clinical investigations for decades. A significant trend was observed consistently in many studies showing that heterozygous complete moles had a higher risk of post-molar GTNs than homozygous complete moles and a few investigations found it statistically significant [60,61,62,63,64,65]. Recently, using DNA genotyping classification, a Japanese study of 27 patients found a significantly higher risk of post-molar gestational trophoblastic disease in heterozygous complete moles than homozygous ones [66]. In our recent study of 1245 consecutive POCs at a major obstetric and gynecological hospital in China [67], we observed that among 165 patients with complete mole, post-molar gestational trophoblastic disease developed in 11.6% (16/165 cases) of homozygous complete moles and in 37% (10/27 cases) of heterozygous complete moles (p = 0.0009). Our study also confirmed previous observations [66] that heterozygous complete moles had a higher average serum hCG level at the initial evacuation than homozygous complete moles (183,100 vs. 119,246 mIU/ml, respectively). Another recent investigation of 204 sporadic hydatidiform moles in Canada also reported a significantly higher risk for post-molar GTN in patients with heterozygous complete moles (91.7%) compared with homozygous ones (48.4%) [68].
Taken together, the overall data conclusively establish that heterozygous/dispermic complete moles are clinically more aggressive with a significantly higher risk for the development of post-molar GTN than homozygous/monospermic complete moles. Therefore, genotyping classification of complete moles based on their genetic zygosity is important for clinical risk assessment for post-molar GTN.
Genotyping in correlation with histomorphology and p57 immunohistochemistry is essential for resolving various diagnostic dilemmas
Molar and non-molar gestations with discrepant/inconsistent p57 expression
P57 immunohistochemical staining is an important ancillary marker for confirmation of a complete molar gestation. P57 is a gene product of CDKN1C on chromosome 11p15.5, a cyclin-dependent kinase inhibitor [69]. The gene is paternally imprinted and the maternal allele is responsible for its nuclear expression in villous cytotrophoblasts and stromal cells [70]. In gestational tissue, loss of the expected p57 expression pattern may occur in the following settings: diandric complete mole, familial biparental complete mole (see below), paternal uniparental disomy of chromosome 11 (see below), loss of maternal p57 allele in PHM [12, 71], and mutations of the maternal allele of p57 gene in a non-molar gestation. Although complete hydatidiform moles characteristically demonstrate loss of p57 expression in villous cytotrophoblast and stromal cells (Fig. 1) as a result of their paternal-only genome, rare CHMs may show normal expression pattern of p57, attributable to a retained maternal chromosome 11 (location of the p57 gene locus) [72, 73]. Moreover, in some complete moles, focal p57 nuclear staining may be seen in 10% or less of the molar villi due to either relaxation of p57 genomic imprinting or technical overstain. Rare partial moles and non-molar hydropic gestations may demonstrate p57 positivity involving only 10–50% of the villi. Aberrant p57 expression has also been documented in Beckwith–Wiedemann syndrome [74].
To further complicate the matter, depending on the presence or absence of maternal genome in different cellular constituents, androgenetic/biparental mosaic gestations may demonstrate “discordant” p57 immunostaining patterns where combination/admixture of negative and positive staining patterns are seen in cytotrophoblast and villous stromal cells within the same villi [4, 75,76,77]. As expected, STR genotyping without isolation of distinct cellular populations for analysis may be difficult to interpret due to variable parental allelic contributions. A “divergent” p57 staining pattern is characterized by two populations of villi; each may have different morphologies and different staining patterns of p57 [46]. This is typically seen in an androgenetic/biparental mosaic gestation with a CHM component (Fig. 6), where the presence of two morphologically distinct villous populations (non-molar villi and molar villi) may result in misdiagnosis of PHM by both histological evaluation and STR genotyping [77]. Twin gestations with one of the twins developing into CHM can easily show two distinct villous populations, one of which has the histological and p57 immunohistochemical features of CHM [4, 54, 55].
The chorionic villi are enlarged, abnormally shaped, and have hypercellular villous stroma (upper left). One villous population demonstrates abnormal trophoblastic proliferation (Villi B, upper right) and loss of p57 nuclear staining in both cytotrophoblast and stromal cells (Villi B, lower left), whereas the other villous population shows retained expression of p57 only in cytotrophoblast (Villi A, lower left) without trophoblastic proliferation (Villi A, upper right). STR genotyping of laser-microdissected cell types shows a biparental allelic pattern in the cytotrophoblast but a homozygous paternal-only pattern in the villous stromal cells (lower right, Endo, normal endometrium; Troph, cytotrophoblast; Strom, villous stromal cells).
In all of the above scenarios of diagnostic difficulty, careful assessment of the histology and p57 staining pattern is important, and DNA genotyping of well-isolated or even laser-microdissected distinct villous/cell populations is needed to reach a definitive diagnosis.
Biparental complete mole mimics normal biparental gestation at genotyping level
Although genotyping provides a precise classification of sporadic moles, one exception is the familial biparental complete mole resulting from inherited NLRP7 or KHDC3L mutations [24, 26, 27, 29, 78]. These biparental complete moles have pathological features indistinguishable from those of the sporadic complete mole. However, molecular genotyping will demonstrate a balanced biparental diploid profile and, therefore, in isolation could be misinterpreted as a normal gestation by STR genotyping. Recognition of this rare type of complete mole requires close correlation of genotyping data with histomorphology and p57 immunohistochemistry. Biparental complete mole exhibits similar histological features to sporadic complete mole and also shows loss of p57 expression in villous cytotrophoblast and stromal cells. In contrast to common sporadic androgenetic complete moles, patients with familial biparental complete mole have an exceedingly rare likelihood of a subsequent normal pregnancy; thus, identification of the condition is clinically important for future fertility planning. Germline sequencing of the NLRP7 and/or KHDC3L gene is required to establish the diagnosis of FBCHM.
Paternal uniparental disomy of chromosome 11 simulating hydatidiform moles
Uniparental disomy is an abnormal genetic condition in which both homolog chromosomes or part of a chromosome are inherited from one parent and the other parent’s homolog chromosome is lost [79]. Uniparental isodisomy occurs when both chromosome copies are homozygous, as a result of duplication of one of the parental chromosomes. Chromosome 11p15.5 region contains a large number of paternally imprinted genes that are important for human placental development and function [80]. It is hypothesized that paternal uniparental disomy of either regional or entire chromosome 11 may lead to altered imprinting gene regulation, sufficient for the development of molar-like conditions. Three cases of paternal uniparental isodisomy at the tyrosine hydroxylase locus on chromosome 11p15.4 were recently identified by STR genotyping with clinical, histological, and p57 immunophenotypical features simulating either partial or complete molar gestations [81]. One case presented with marked villous enlargement, two villous populations, and hydrops with cistern formation (Fig. 7) with a normal p57 staining pattern, simulating a PHM. Another case demonstrated histological features reminiscent of an early complete mole, including abnormal villous configuration, diffuse villous hydrops, abnormal trophoblastic hyperplasia, hypercellular villous stroma with frequent karyorrhexis, and the absence of fetal red blood cells. Paradoxically, p57 immunostain showed normal expression in this case. The third case was remarkable for marked atypical villous trophoblastic proliferation with abnormal loss of p57 staining, but without other histological features of either partial or complete mole (Fig. 7). This patient developed clinical complications simulating persistent GTN with rising serum hCG several weeks after the initial evacuation, for which she initially received single-agent chemotherapy. Her disease progressed with uterine and lung mass lesions, eventually requiring multiagent chemotherapy, ultimately leading to normalization of serum hCG and a long-term remission. Careful genotyping analysis with attention to the STR loci at the chromosome 11p15.5 region is needed to recognize this rare entity and patients with missed abortion due to paternal uniparental disomy of chromosome 11, in particular those with significant atypical trophoblastic proliferation and abnormal p57 expression, should be followed clinically according to the post-molar serum hCG surveillance program.
Hydropic changes with cistern formation and normal p57 expression are seen in one case (upper left and right). Irregularly shaped chorionic villi with abnormal trophoblastic proliferation and loss of p57 expression in cytotrophoblast and villous stromal cells are seen in another case (lower left and right). Note the presence of positive internal control in intervillous intermediate trophoblast in the lower right corner of the image.
Egg-donor gestation mimics complete mole at genotyping level
Abortion specimens from an egg-donor gestation may present a unique challenge for genotyping diagnosis of molar gestation. Microscopic examination may reveal mildly hydropic, dysmorphic chorionic villi, with occasional trophoblastic pseudo-inclusions, raising concern for a PHM and prompting DNA genotyping confirmation. As the gestation contains the genome from the egg donor and does not inherit the recipient mother’s genome, DNA genotyping of chorionic villous tissue will demonstrate a distinct allelic pattern—not present in the recipient’s decidual tissue. Without awareness of the clinical history of donor pregnancy, this can be mistaken for a diandric, paternal-only genome, leading to an erroneous molecular interpretation of dispermic/heterozygous complete mole [82, 83]. Therefore, if the histological features are only suspicious for PHM, not for CHM, careful correlation of the genotyping data with histomorphology and p57 immunohistochemistry is crucial, and clinical inquiry about the history of egg-donor pregnancy is confirmatory.
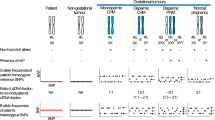
Algorithmic approach in the diagnostic workup of hydatidiform moles
Given the significant limitations of histomorphology alone in the diagnosis of molar gestations, algorithmic approaches combining morphology, p57 immunohistochemistry, and DNA genotyping have been advocated in recent years [4, 12, 16, 46]. Once a suspicion is made on the initial histological evaluation, three possible algorithms may be considered, depending on the type of mole in question and the availability of ancillary tools (Fig. 8). The most efficient and precise algorithm is to subject all suspected specimens to genotyping as a “one-stop shopping” approach and p57 immunohistochemistry is only used to address any discordance between morphology and genotyping to identify biparental CHM, mosaicism/chimerism, and hydatidiform mole involving a twin gestation. The second approach is to triage specimens suspected for CHM to p57 immunohistochemistry for confirmation, and those suspected for PHM to STR genotyping. The third approach is to subject all suspected molar cases to p57 immunostain to identify CHM and genotyping is only applied to the remaining to distinguish PHM from non-molar gestations. Given the fact that genetic subclassification of CHM as homo- vs. heterozygous provides an important prognostic value for clinical management, the first “one-stop shopping” approach should be considered if STR genotyping is readily available. In the setting of limited resources, use of ancillary techniques may be focused on identifying CHM due to its greatest risk for post-molar GTN by applying p57 immunohistochemistry and foregoing genotyping to separate PHM from non-molar gestations with an understanding that some gestations may be underdiagnosed, while others may be over-diagnosed as PHM leading to unnecessary patient follow-up and contraception.
Products of conceptions with morphological suspicion for hydatidiform moles can be uniformly subjected to STR genotyping as a “one-stop shopping” approach (A). The second approach is to perform either STR genotyping or p57 immunohistochemistry based on the histological suspicion for either CHM or PHM, respectively (B). The third approach is to submit all suspected specimens for p57 immunohistochemistry to confirm CHM and then triage those with normal p57 expression for STR genotyping to distinguish PHM from non-molar gestations (C).
DNA genotyping for separating gestational trophoblastic tumors from non-gestational tumors with overlapping histology
As a result of successful clinical post-molar surveillance program, GTN is nowadays diagnosed primarily based on serum hCG measurement combined with imaging studies. Thus, chemotherapeutic treatment of patients with GTN is generally initiated without a tissue/pathological confirmation and post-molar gestational trophoblastic tumors are much less often encountered in a pathology lab. Gestational choriocarcinoma is the most common GTN that typically develops in the reproductive age (average of 30 years old), following a normal gestation, hydatidiform mole, or abortion in 50%, 22.5%, and 20% of the cases, respectively [84, 85]. Although the most common clinical symptom is vaginal bleeding, the first presentation may be extrauterine hemorrhage as a result of metastasis [86] to extrauterine sites including the vagina, lung, liver, brain, kidney, and abdomen [87, 88]. While the time interval between choriocarcinoma and the antecedent gestation is usually 1–3 months on average after a term pregnancy, the time interval is 13 months on average following a complete mole, but rarely it may be over 20 years [89, 90]. Gestational choriocarcinoma involving the fallopian tube or ovary may arise from an ectopic pregnancy [91]. In rare cases of extrauterine presentation of gestational choriocarcinoma arising in a term placenta, the primary intraplacental choriocarcinoma may be occult and only identified retrospectively by thorough gross and microscopic examination of the term placenta [92, 93]. Therefore, gestational choriocarcinomas presenting at an extrauterine site without a recently documented history of GTD may pose a significant diagnostic challenge, not only because they can be mistaken as somatic carcinomas but also because tumors with trophoblastic differentiation can have different cellular origins other than gestational.
Non-gestational choriocarcinoma of germ cell origin in female patients usually occurs in children or in young adults, involves the ovary, and may contain other non-choriocarcinomatous components as part of a mixed germ cell tumor [94]. Somatic carcinomas at various primary sites may also show trophoblastic differentiation—in the form of a choriocarcinomatous morphology or as scattered syncytiotrophoblastic giant cells, mimicking metastatic gestational choriocarcinoma. Helpful clinical features in favor of somatic origin include older patient age, postmenopausal status, and a relatively lower level of serum beta-hCG (usually < 10,000 mIU/mL) [95, 96]. However, patient age, menstrual status, pregnancy history, and tumor location are not necessarily reliable for determining the gestational vs. non-gestational nature of a choriocarcinoma.
Choriocarcinoma typically forms bulky, destructive mass lesions with extensive hemorrhage and necrosis within the involved organ. At the histological level, regardless of its pathogenetic origin, the tumor consists of bi- or triphasic arrangements of mononuclear trophoblastic cells and multinucleated syncytiotrophoblast. The nuclear atypia is marked, often with bizarre nuclei and there is brisk mitotic activity with frequent atypical mitotic figures. Abundant tumor necrosis and hemorrhage are present. Germ cell choriocarcinomas and somatic carcinomas with trophoblastic differentiation typically also contain recognizable other histologic components: other germ cell tumor types (yolk sac tumor, dysgerminoma, and immature teratoma) or somatic carcinoma components (adenocarcinoma, clear cell carcinoma, squamous cell carcinoma, giant cell carcinoma, or urothelial carcinoma), respectively [95,96,97,98,99]. However, germ cell choriocarcinoma of pure histology can occur and the conventional carcinoma components in a somatic carcinoma may require thorough sampling, and may not always be present in a small biopsy specimen. Trophoblastic immunohistochemical markers including hCG, hPL, GATA3, and inhibin are helpful in confirming trophoblastic differentiation, but do not differentiate between the different pathogenetic entities [100, 101]. SALL4 is expressed in choriocarcinoma of both germ cell and gestational origin [102, 103].
Gestational choriocarcinoma is highly chemosensitive and responds well to single-agent methotrexate (low-risk disease) or EMA-CO (etoposide, methotrexate, actinomycin D, cyclophosphamide, vincristine) combination chemotherapy (high-risk disease) with an excellent prognosis [84]. Germ cell choriocarcinoma requires surgical staging followed by bleomycin-based chemotherapy. Somatic carcinomas with trophoblastic differentiation, on the other hand, typically have an aggressive clinical course with poor response to chemotherapy [104]. Genotyping detection of a distinct paternal genetic complement not present in the patient’s normal tissues definitively separates a gestational trophoblastic tumor from a non-gestational neoplasm of either germ cell or somatic nature. In addition, genotyping also allows for precise identification of the index causative gestational event (hydatidiform mole, abortion, or term pregnancy) of gestational choriocarcinoma [90, 105,106,107,108]. Our recent study of three young women with initial presentation of lung tumors with choriocarcinoma morphology highlights some of the most significant diagnostic challenges [109]. Of the three patients (ages 37–48 years), two were noted to have elevated serum β-hCG levels at the time of their presentation, whereas serum β-hCG was not evaluated preoperatively in the third patient. None of them had a clinical history of molar pregnancy or GTN. Core biopsies of the lung masses were performed in two patients and one patient underwent a wedge resection, showing high-grade carcinoma in all three cases. β-hCG immunostain was performed in two cases and showed diffuse immunoreactivity. Clinical history and imaging studies were not conclusive in any of the cases to rule out a gestational origin. Ultimately, STR genotyping analysis was performed to compare the allelic differences between tumor and normal tissues, revealing an identical profile in one case consistent with primary lung carcinoma, and distinct paternal alleles confirming metastatic gestational choriocarcinoma in the other two patients (Fig. 9), both of whom responded well to combined chemotherapy. In contrast, the patient with primary lung carcinoma experienced rapid progression with liver, bone, and brain metastases despite combination chemotherapy and died 15 months after the diagnosis. Presentation of a high-grade carcinoma at an extrauterine site in a young woman poses a unique diagnostic challenge in the absence of history of GTD. High index of suspicion is crucial and a differential diagnosis of metastatic gestational trophoblastic tumor must be considered. Molecular genotyping is a powerful tool to separate a malignant somatic or germ cell tumor with trophoblastic differentiation from a metastatic gestational trophoblastic tumor, including gestational choriocarcinoma, placental site trophoblastic tumor (PSTT), and epithelioid trophoblastic tumor (ETT) [105, 110,111,112] (Fig. 10).
A large necrotic lung mass was the initial presentation (left) with characteristic biphasic proliferation of markedly atypical trophoblast (right upper). STR genotyping shows the presence of distinct paternal alleles (asterisks) in the tumor at multiple STR loci, confirming its gestational origin (right lower).
Comparison of the allelic patterns at STR loci between the tumor and paired normal tissue identifies the presence or absence of paternal alleles in the tumor, allowing for diagnostic separation of a gestational tumor from its non-gestational mimics. Genotyping also allows precise recognition of the index gestational event (hydatidiform mole, abortion, or term pregnancy) of gestational trophoblastic tumors.
Genotyping provides crucial data for FIGO/WHO risk scoring of gestational trophoblastic tumors
Once a diagnosis of gestational trophoblastic tumor is made, subsequent clinicopathological assessment by the FIGO/WHO risk scoring scheme is performed to triage the patient into low- or high-risk category for single-agent or combination chemotherapy, respectivley [113]. The following clinicopathological parameters are included in the risk score: patient age, type of index pregnancy, interval months from the index pregnancy, pretreatment serum hCG, size and number of metastases, and previous failed chemotherapy [114]. Gestational choriocarcinoma arising from a molar pregnancy is of lower risk than that related to a non-molar abortion or a term pregnancy, and a shorter time interval since the index pregnancy is a more favorable factor. However, in the absence of tissue diagnosis in many GTNs, the type of antecedent pregnancy and the time interval from the index pregnancy may be uncertain in some cases [84], as the immediate antecedent pregnancy may not be the index/causative gestation in a patient with history of multiple pregnancies. When in doubt, molecular genotyping of the choriocarcinoma in comparison with the suspected gestation(s) may identify the true causative gestational event, and therefore the time interval between the tumor and its index gestation can be accurately calculated [4].
It is important to note that although there are four trophoblastic tumor types listed under GTN, invasive mole and choriocarcinoma are by far the most common and they follow the FIGO/WHO risk scoring system for triaging the patient for subsequent chemotherapy [114]. However, PSTT and ETT are different clinical entities, as they are mostly diagnosed months or years after their index gestation and are generally not part of the post-molar GTN spectrum and the FIGO/WHO risk scoring system is not appropriate. Patients with PSTT and ETT require hysterectomy with or without salpingo-oophorectomy for management and, depending on the pathological stage, they may or may not receive adjuvant chemotherapy. Although a patient with PSTT or ETT does not need genotyping for FIGO/WHO risk scoring, genotyping may be used to confirm a diagnosis of PSTT and ETT in separating them from somatic tumors (in particular squamous cell carcinoma and epithelioid leiomyosarcoma). However, genotyping has no role in the diagnostic separation between various gestational trophoblastic tumors (ETT vs. PSTT vs. gestational choriocarcinoma) and their reactive or preneoplastic counterparts (PSTT vs. exaggerated placental site (EPS) or ETT vs. atypical placental site nodule (APSN) vs. placental site nodule (PSN)) [112, 115]. Furthermore, genotyping analysis does not have a prognostic value in this setting.
Conclusions
In recent decades, applications of STR genotyping have revolutionized our ability to precisely diagnose and subclassify various gestational trophoblastic disease entities, essential for the optimal clinical management of the patient. Such molecular integration extends beyond the diagnosis and clinical patient care, as we are now able to investigate the epidemiology, pathogenesis, and biological behavior of gestational trophoblastic disease in an unprecedented manner of accuracy, largely because precise study cohorts of the disease entity can be secured as a result of DNA genotyping.
References
Froeling FE, Seckl MJ. Gestational trophoblastic tumours: an update for 2014. Curr Oncol Rep. 2014;16:408.
Brown J, Naumann RW, Seckl MJ, Schink J. 15years of progress in gestational trophoblastic disease: scoring, standardization, and salvage. Gynecol Oncol. 2017;144:200–7.
Fukunaga M, et al. Interobserver and intraobserver variability in the diagnosis of hydatidiform mole. Am J Surg Pathol. 2005;29:942–7.
Hui P, Buza N, Murphy KM, Ronnett BM. Hydatidiform moles: genetic basis and precision diagnosis. Annu Rev Pathol. 2017;12:449–85.
Hancock, BW, Newlands, ES, Berkowitz, RS. Gestational trophoblastic disease 1st edn. London; New York: Chapman & Hall Medical; 1997.
Curry SL, Hammond CB, Tyrey L, Creasman WT, Parker RT. Hydatidiform mole: diagnosis, management, and long-term followup of 347 patients. Obstet Gynecol. 1975;45:1–8.
Kohorn EI. Hydatidiform mole and gestational trophoblastic disease in Southern Connecticut. Obstet Gynecol. 1982;59:78–84.
Lurain JR, Brewer JI, Torok EE, Halpern B. Natural history of hydatidiform mole after primary evacuation. Am J Obstet Gynecol. 1983;145:591–5.
Rice LW, Berkowitz RS, Lage JM, Goldstein DP, Bernstein MR. Persistent gestational trophoblastic tumor after partial hydatidiform mole. Gynecol Oncol. 1990;36:358–62.
Gaber LW, Redline RW, Mostoufi-Zadeh M, Driscoll SG. Invasive partial mole. Am J Clin Pathol. 1986;85:722–4.
Seckl MJ, et al. Choriocarcinoma and partial hydatidiform moles. Lancet. 2000;356:36–39.
Banet N, et al. Characteristics of hydatidiform moles: analysis of a prospective series with p57 immunohistochemistry and molecular genotyping. Mod Pathol. 2014;27:238–54.
Vang R, et al. Diagnostic reproducibility of hydatidiform moles: ancillary techniques (p57 immunohistochemistry and molecular genotyping) improve morphologic diagnosis. Am J Surg Pathol. 2012;36:443–53.
Furtado LV, et al. Diagnostic utility of microsatellite genotyping for molar pregnancy testing. Arch Pathol Lab Med. 2013;137:55–63.
Colgan TJ, Chang MC, Nanji S, Kolomietz E. DNA genotyping of suspected partial hydatidiform moles detects clinically significant aneuploidy. Int J Gynecol Pathol. 2017;36:217–21.
Buza N, Hui P. Immunohistochemistry and other ancillary techniques in the diagnosis of gestational trophoblastic diseases. Semin Diagn Pathol. 2014;31:223–32.
Kajii T, Ohama K. Androgenetic origin of hydatidiform mole. Nature. 1977;268:633–4.
Szulman AE. Syndromes of hydatidiform moles. Partial vs. complete. J Reprod Med. 1984;29:788–91.
Wallace DC, Surti U, Adams CW, Szulman AE. Complete moles have paternal chromosomes but maternal mitochondrial DNA. Hum Genet. 1982;61:145–7.
Azuma C, et al. Application of gene amplification by polymerase chain reaction to genetic analysis of molar mitochondrial DNA: the detection of anuclear empty ovum as the cause of complete mole. Gynecol Oncol. 1991;40:29–33.
Jacobs PA, Wilson CM, Sprenkle JA, Rosenshein NB, Migeon BR. Mechanism of origin of complete hydatidiform moles. Nature. 1980;286:714–6.
Ambani LM, Vaidya RA, Rao CS, Daftary SD, Motashaw ND. Familial occurrence of trophoblastic disease - report of recurrent molar pregnancies in sisters in three families. Clin Genet. 1980;18:27–29.
Williams D, Hodgetts V, Gupta J. Recurrent hydatidiform moles. Eur J Obstet Gynecol Reprod Biol. 2010;150:3–7.
Murdoch S, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38:300–2.
Qian J, Deveault C, Bagga R, Xie X, Slim R. Women heterozygous for NALP7/NLRP7 mutations are at risk for reproductive wastage: report of two novel mutations. Hum Mutat. 2007;28:741.
Kou YC, et al. A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod. 2008;14:33–40.
Wang CM, et al. Identification of 13 novel NLRP7 mutations in 20 families with recurrent hydatidiform mole; missense mutations cluster in the leucine-rich region. J Med Genet. 2009;46:569–75.
Nguyen NM, Slim R. Genetics and epigenetics of recurrent hydatidiform moles: basic science and genetic counselling. Curr Obstet Gynecol Rep. 2014;3:55–64.
Parry DA, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011;89:451–8.
Reddy R, et al. Report of four new patients with protein-truncating mutations in C6orf221/KHDC3L and colocalization with NLRP7. Eur J Hum Genet. 2013;21:957–64.
Genest DR, et al. Do nontriploid partial hydatidiform moles exist? A histologic and flow cytometric reevaluation of nontriploid specimens. J Reprod Med. 2002;47:363–8.
Lawler SD, Fisher RA, Pickthall VJ, Povey S, Evans MW. Genetic studies on hydatidiform moles. I. The origin of partial moles. Cancer Genet Cytogenet. 1982;5:309–20.
Jacobs PA, Szulman AE, Funkhouser J, Matsuura JS, Wilson CC. Human triploidy: relationship between parental origin of the additional haploid complement and development of partial hydatidiform mole. Ann Hum Genet. 1982;46:223–31.
Surti U, Szulman AE, Wagner K, Leppert M, O’Brien SJ. Tetraploid partial hydatidiform moles: two cases with a triple paternal contribution and a 92,XXXY karyotype. Hum Genet. 1986;72:15–21.
Sheppard DM, Fisher RA, Lawler SD, Povey S. Tetraploid conceptus with three paternal contributions. Hum Genet. 1982;62:371–4.
Murphy KM, et al. Tetraploid partial hydatidiform mole: a case report and review of the literature. Int J Gynecol Pathol. 2012;31:73–79.
Zhivotovsky LA, Bennett L, Bowcock AM, Feldman MW. Human population expansion and microsatellite variation. Mol Biol Evol. 2000;17:757–67.
Collins PJ, et al. Developmental validation of a single-tube amplification of the 13 CODIS STR loci, D2S1338, D19S433, and amelogenin: the AmpFlSTR Identifiler PCR Amplification Kit. J Forensic Sci. 2004;49:1265–77.
Hui P. Molecular diagnosis of gestational trophoblastic disease. Expert Rev Mol Diagn. 2010;10:1023–34.
Baine I, Hui P. Practical applications of DNA genotyping in diagnostic pathology. Expert Rev Mol Diagn. 2019;19:175–88.
Lipata F, et al. Precise DNA genotyping diagnosis of hydatidiform mole. Obstet Gynecol. 2010;115:784–94.
Bifulco C, et al. Genotypic analysis of hydatidiform mole: an accurate and practical method of diagnosis. Am J Surg Pathol. 2008;32:445–51.
Chew SH, Perlman EJ, Williams R, Kurman RJ, Ronnett BM. Morphology and DNA content analysis in the evaluation of first trimester placentas for partial hydatidiform mole (PHM). Hum Pathol. 2000;31:914–24.
Norris-Kirby A, Hagenkord JM, Kshirsagar MP, Ronnett BM, Murphy KM. Abnormal villous morphology associated with triple trisomy of paternal origin. J Mol Diagn. 2010;12:525–9.
Redline RW, Hassold T, Zaragoza M. Determinants of villous trophoblastic hyperplasia in spontaneous abortions. Mod Pathol. 1998;11:762–8.
Ronnett BM, DeScipio C, Murphy KM. Hydatidiform moles: ancillary techniques to refine diagnosis. Int J Gynecol Pathol. 2011;30:101–16.
WHO. WHO classification of tumours series. 5th edn, Vol. 5th. Lyon, France: International Agnecy for Researcho on Cancer - World Health Organization; 2020. p. 317–8.
Buza N, Hui P. Partial hydatidiform mole: histologic parameters in correlation with DNA genotyping. Int J Gynecol Pathol. 2013;32:307–15.
van Lijnschoten G, Arends JW, Geraedts JP. Comparison of histological features in early spontaneous and induced trisomic abortions. Placenta. 1994;15:765–73.
Zaragoza MV, et al. Parental origin and phenotype of triploidy in spontaneous abortions: predominance of diandry and association with the partial hydatidiform mole. Am J Hum Genet. 2000;66:1807–20.
McFadden DE, Robinson WP. Phenotype of triploid embryos. J Med Genet. 2006;43:609–12.
Hassold T, et al. A cytogenetic study of 1000 spontaneous abortions. Ann Hum Genet. 1980;44:151–78.
Szulman AE, Philippe E, Boue JG, Boue A. Human triploidy: association with partial hydatidiform moles and nonmolar conceptuses. Hum Pathol. 1981;12:1016–21.
Genest DR. Partial hydatidiform mole: clinicopathological features, differential diagnosis, ploidy and molecular studies, and gold standards for diagnosis. Int J Gynecol Pathol. 2001;20:315–22.
Paradinas FJ, Fisher RA, Browne P, Newlands ES. Diploid hydatidiform moles with fetal red blood cells in molar villi. 1–Pathology, incidence, and prognosis. J Pathol. 1997;181:183–8.
Sebire NJ, Fisher RA, Rees HC. Histopathological diagnosis of partial and complete hydatidiform mole in the first trimester of pregnancy. Pediatr Dev Pathol. 2003;6:69–77.
Fukunaga M. Histopathologic study of partial hydatidiform moles and DNA triploid placentas. Pathol Int. 1994;44:528–34.
Jeffers MD, O’Dwyer P, Curran B, Leader M, Gillan JE. Partial hydatidiform mole: a common but underdiagnosed condition. A 3-year retrospective clinicopathological and DNA flow cytometric analysis. Int J Gynecol Pathol. 1993;12:315–23.
Esteban JM, et al. Effects of various fixatives and fixation conditions on DNA ploidy analysis. A need for strict internal DNA standards. Am J Clin Pathol. 1991;95:460–6.
Wake N, et al. Malignant potential of homozygous and heterozygous complete moles. Cancer Res. 1984;44:1226–30.
Wake N, et al. The propensity to malignancy of dispermic heterozygous moles. Placenta. 1987;8:319–26.
Lawler SD, Fisher RA. Genetic studies in hydatidiform mole with clinical correlations. Placenta. 1987;8:77–88.
Lawler SD, Fisher RA, Dent J. A prospective genetic study of complete and partial hydatidiform moles. Am J Obstet Gynecol. 1991;164:1270–7.
Cho S, Kim SJ. Genetic study of hydatidiform moles by restriction fragment length polymorphisms (RFLPs) analysis. J Korean Med Sci. 1993;8:446–52.
Niemann I, Hansen ES, Sunde L. The risk of persistent trophoblastic disease after hydatidiform mole classified by morphology and ploidy. Gynecol Oncol. 2007;104:411–5.
Baasanjav B, et al. The risk of post-molar gestational trophoblastic neoplasia is higher in heterozygous than in homozygous complete hydatidiform moles. Hum Reprod. 2010;25:1183–91.
Zheng XZ, et al. Heterozygous/dispermic complete mole confers a significantly higher risk for post-molar gestational trophoblastic disease. Mod Pathol. 2020;33:1979–88.
Khawajkie Y, et al. Correction: Comprehensive analysis of 204 sporadic hydatidiform moles: revisiting risk factors and their correlations with the molar genotypes. Mod Pathol. 2020;33:1237.
Chilosi M, et al. Differential expression of p57kip2, a maternally imprinted cdk inhibitor, in normal human placenta and gestational trophoblastic disease. Lab Invest. 1998;78:269–76.
Fukunaga M. Immunohistochemical characterization of p57(KIP2) expression in early hydatidiform moles. Hum Pathol. 2002;33:1188–92.
DeScipio C, et al. Diandric triploid hydatidiform mole with loss of maternal chromosome 11. Am J Surg Pathol. 2011;35:1586–91.
Fisher RA, et al. Complete hydatidiform mole retaining a chromosome 11 of maternal origin: molecular genetic analysis of a case. Mod Pathol. 2004;17:1155–60.
McConnell TG, Norris-Kirby A, Hagenkord JM, Ronnett BM, Murphy KM. Complete hydatidiform mole with retained maternal chromosomes 6 and 11. Am J Surg Pathol. 2009;33:1409–15.
Ronnett BM. Hydatidiform moles: ancillary techniques to refine diagnosis. Arch Pathol Lab Med. 2018;142:1485–502.
Hoffner L, Dunn J, Esposito N, Macpherson T, Surti U. P57KIP2 immunostaining and molecular cytogenetics: combined approach aids in diagnosis of morphologically challenging cases with molar phenotype and in detecting androgenetic cell lines in mosaic/chimeric conceptions. Hum Pathol. 2008;39:63–72.
Hoffner L, Parks WT, Swerdlow SH, Carson JC, Surti U. Simultaneous detection of imprinted gene expression (p57(KIP2)) and molecular cytogenetics (FICTION) in the evaluation of molar pregnancies. J Reprod Med. 2010;55:219–28.
Lewis GH, et al. Characterization of androgenetic/biparental mosaic/chimeric conceptions, including those with a molar component: morphology, p57 immnohistochemistry, molecular genotyping, and risk of persistent gestational trophoblastic disease. Int J Gynecol Pathol. 2013;32:199–214.
Van den Veyver IB, Al-Hussaini TK. Biparental hydatidiform moles: a maternal effect mutation affecting imprinting in the offspring. Hum Reprod Update. 2006;12:233–42.
Sebire NJ, May PC, Kaur B, Seckl MJ, Fisher RA. Abnormal villous morphology mimicking a hydatidiform mole associated with paternal trisomy of chromosomes 3,7,8 and unipaternal disomy of chromosome 11. Diagn Pathol. 2016;11:20.
Hui, P. Gestational trophoblastic disease: diagnostic and molecular genetic pathology. New York: Springer; 2012.
Buza N, McGregor SM, Barroilhet L, Zheng X, Hui P. Paternal uniparental isodisomy of tyrosine hydroxylase locus at chromosome 11p15.4: spectrum of phenotypical presentations simulating hydatidiform moles. Mod Pathol. 2019;32:1180–8.
Joseph NM, Pineda C, Rabban JT. DNA genotyping of nonmolar donor egg pregnancies with abnormal villous morphology: allele zygosity patterns prevent misinterpretation as complete hydatidiform mole. Int J Gynecol Pathol. 2018;37:191–7.
Buza N, Hui P. Egg donor pregnancy: a potential pitfall in DNA genotyping diagnosis of hydatidiform moles. Int J Gynecol Pathol. 2014;33:507–10.
Lurain JR. Gestational trophoblastic disease II: classification and management of gestational trophoblastic neoplasia. Am J Obstet Gynecol. 2011;204:11–18.
Hui, P. Gestational choriocarcinoma. In: Gestational trophoblastic disease. Current Clinical Pathology. New York: Springer; 2012. p. 127–37. https://doi.org/10.1007/978-1-61779-394-3_8.
Horn LC, Bilek K. Clinicopathologic analysis of gestational trophoblastic disease–report of 158 cases. Gen Diagn Pathol. 1997;143:173–8.
Ober WB, Edgcomb JH, Price EB Jr. The pathology of choriocarcinoma. Ann NY Acad Sci. 1971;172:299–426.
Morgan JM, Lurain JR. Gestational trophoblastic neoplasia: an update. Curr Oncol Rep. 2008;10:497–504.
O’Neill CJ, Houghton F, Clarke J, McCluggage WG. Uterine gestational choriocarcinoma developing after a long latent period in a postmenopausal woman: the value of DNA polymorphism studies. Int J Surg Pathol. 2008;16:226–9.
Fisher RA, et al. The impact of molecular genetic diagnosis on the management of women with hCG-producing malignancies. Gynecol Oncol. 2007;107:413–9.
Ober WB, Maier RC. Gestational choriocarcinoma of the fallopian tube. Diagn Gynecol Obstet. 1981;3:213–31.
Liu J, Guo L. Intraplacental choriocarcinoma in a term placenta with both maternal and infantile metastases: a case report and review of the literature. Gynecol Oncol. 2006;103:1147–51.
Ganapathi KA, et al. Incidental finding of placental choriocarcinoma after an uncomplicated term pregnancy: a case report with review of the literature. Int J Gynecol Pathol. 2010;29:476–8.
Kong B, Tian YJ, Zhu WW, Qin YJ. A pure nongestational ovarian choriocarcinoma in a 10-year-old girl: case report and literature review. J Obstet Gynaecol Res. 2009;35:574–8.
Rawish KR, Buza N, Zheng W, Fadare O. Endometrial carcinoma with trophoblastic components: clinicopathologic analysis of a rare entity. Int J Gynecol Pathol. 2018;37:174–90.
Hu YJ, Ip PP, Chan KK, Tam KF, Ngan HY. Ovarian clear cell carcinoma with choriocarcinomatous differentiation: report of a rare and aggressive tumor. Int J Gynecol Pathol. 2010;29:539–45.
Oliva E, Andrada E, Pezzica E, Prat J. Ovarian carcinomas with choriocarcinomatous differentiation. Cancer. 1993;72:2441–6.
Mukonoweshuro P, McCluggage WG. Clear cell carcinoma of the cervix with choriocarcinomatous differentiation: report of an extremely rare phenomenon associated with mismatch repair protein abnormality. Int J Gynecol Pathol. 2017;36:323–7.
Attanoos RL, et al. Pulmonary giant cell carcinoma: pathological entity or morphological phenotype? Histopathology. 1998;32:225–31.
Banet N, et al. GATA-3 expression in trophoblastic tissues: an immunohistochemical study of 445 cases, including diagnostic utility. Am J Surg Pathol. 2015;39:101–8.
Shih IM, Kurman RJ. Immunohistochemical localization of inhibin-alpha in the placenta and gestational trophoblastic lesions. Int J Gynecol Pathol. 1999;18:144–50.
Miettinen M, et al. SALL4 expression in germ cell and non-germ cell tumors: a systematic immunohistochemical study of 3215 cases. Am J Surg Pathol. 2014;38:410–20.
Stichelbout M, et al. SALL4 expression in gestational trophoblastic tumors: a useful tool to distinguish choriocarcinoma from placental site trophoblastic tumor and epithelioid trophoblastic tumor. Hum Pathol. 2016;54:121–6.
Weissferdt A, Moran CA. Primary giant cell carcinomas of the lung: a clinicopathological and immunohistochemical analysis of seven cases. Histopathology. 2016;68:680–5.
Zhao J, et al. Molecular genetic analyses of choriocarcinoma. Placenta. 2009;30:816–20.
Jiao LZ, et al. Clinical analysis of 21 cases of nongestational ovarian choriocarcinoma. Int J Gynecol Cancer. 2010;20:299–302.
Shahib N, et al. Genetic origin of malignant trophoblastic neoplasms analyzed by sequence tag site polymorphic markers. Gynecol Oncol. 2001;81:247–53.
Arima T, et al. Malignant trophoblastic neoplasms with different modes of origin. Cancer Genet Cytogenet. 1995;85:5–15.
Buza N, Baine I, Hui P. Precision genotyping diagnosis of lung tumors with trophoblastic morphology in young women. Mod Pathol. 2019;32:1271–80.
Buza N, Rutherford T, Hui P. Genotyping diagnosis of nongestational choriocarcinoma involving fallopian tube and broad ligament: a case study. Int J Gynecol Pathol. 2014;33:58–63.
Savage J, Adams E, Veras E, Murphy KM, Ronnett BM. Choriocarcinoma in women: analysis of a case series with genotyping. Am J Surg Pathol. 2017;41:1593–606.
Xu ML, Yang B, Carcangiu ML, Hui P. Epithelioid trophoblastic tumor: comparative genomic hybridization and diagnostic DNA genotyping. Mod Pathol. 2009;22:232–8.
Kohorn EI. Negotiating a staging and risk factor scoring system for gestational trophoblastic neoplasia. A progress report. J Reprod Med. 2002;47:445–50.
Ngan HYS, et al. Update on the diagnosis and management of gestational trophoblastic disease. Int J Gynaecol Obstet. 2018;143:79–85.
Fadare O, Parkash V, Carcangiu ML, Hui P. Epithelioid trophoblastic tumor: clinicopathological features with an emphasis on uterine cervical involvement. Mod Pathol. 2006;19:75–82.
Funding
No funding is involved in preparing the manuscript.
Author information
Authors and Affiliations
Contributions
Both authors contributed to the writing of the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Buza, N., Hui, P. Genotyping diagnosis of gestational trophoblastic disease: frontiers in precision medicine. Mod Pathol 34, 1658–1672 (2021). https://doi.org/10.1038/s41379-021-00831-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00831-9
This article is cited by
-
Practical guidelines of the EOTTD for pathological and genetic diagnosis of hydatidiform moles
Virchows Archiv (2024)
-
When a vesicular placenta meets a live fetus: case report of twin pregnancy with a partial hydatidiform mole
BMC Pregnancy and Childbirth (2021)