Abstract
Well-differentiated hepatocellular neoplasms are currently classified in the World Health Organization scheme as hepatocellular adenoma or hepatocellular carcinoma. There is no recognized diagnostic category for atypical cases with borderline features, and we have designated these as atypical hepatocellular neoplasms. Diffuse glutamine synthetase staining is used as a surrogate marker to detect β-catenin activation, a well-recognized high risk feature in hepatocellular tumors. This study examined 27 well-differentiated hepatocellular neoplasms with diffuse glutamine synthetase staining, including 7 atypical hepatocellular neoplasms with no cytoarchitectural atypia, 6 atypical hepatocellular neoplasms with focal cytoarchitectural atypia, and 14 well-differentiated hepatocellular carcinomas. Capture-based next-generation sequencing was performed, and alterations in WNT pathway genes (CTNNB1, APC, AXIN1) were seen in 81% of cases (10/13 atypical hepatocellular neoplasms and 12/14 of hepatocellular carcinomas), while the molecular basis of diffuse glutamine synthetase staining was unclear in the remaining 19% of cases. Additional non-WNT pathway mutations (TP53, TSC1, DNMT3A, CREBBP) or copy number alterations were present in 56% of atypical hepatocellular neoplasms, with no significant difference in cases with or without focal cytoarchitectural atypia, supporting that all cases with β-catenin activation should be classified as atypical irrespective of atypia. Atypical hepatocellular neoplasm and hepatocellular carcinoma also demonstrated largely similar genomic profiles, but TERT promoter mutations were restricted to hepatocellular carcinoma (21%) and copy number alterations were more common in hepatocellular carcinoma (64 vs 31%). Mutational and copy number analysis may be helpful in characterization and risk stratification of atypical hepatocellular neoplasms when morphology and glutamine synthetase staining yield ambiguous results.
Similar content being viewed by others
Introduction
Activation of β-catenin is a well-recognized high risk feature in liver tumors with morphologic features of hepatocellular adenoma [1,2,3,4,5]. These tumors are categorized as β-catenin-activated hepatocellular adenomas in the World Health Organization 2010 classification [6]. Compared to other hepatocellular adenoma subtypes, β-catenin-activated tumors are frequently associated with male gender, advanced age and hepatocellular carcinoma at diagnosis or on follow-up [1, 2, 7]. Atypical cytoarchitectural features, including cytologic atypia, pseudoacinar architecture, focally thickened cell plates, small cell change and reticulin loss occur in 40–80% of these tumors [2, 3, 5]. These changes are often focal and not sufficient for an unequivocal diagnosis of hepatocellular carcinoma. Cytogenetic changes typical of hepatocellular carcinoma such as gains of chromosomes 1 and 8 have been described in these tumors [2, 8, 9]. Due to the presence of atypical pathologic/cytogenetic features and frequent association with hepatocellular carcinoma, it has been advocated that these tumors should not be classified as a subtype of hepatocellular adenoma and are more appropriately designated as atypical hepatocellular neoplasms [2]. Other terms like hepatocellular neoplasm of uncertain malignant potential (HUMP) and borderline lesions have also been used [4, 9,10,11,12].
In view of the high risk of hepatocellular carcinoma in hepatocellular adenoma -like tumors with β-catenin activation, resection of these tumors is recommended [7, 13]. This puts the onus on the pathologist to accurately determine β-catenin activation in these tumors to facilitate proper management. β-catenin activation most commonly results from mutations or deletions involving exon 3 of the CTNNB1 (β-catenin) gene leading to nuclear translocation of β-catenin, which can be demonstrated by immunohistochemistry [1, 3, 4, 14]. Nuclear β-catenin leads to transcriptional activation of glutamine synthetase. In normal liver, glutamine synthetase is confined to a few hepatocytes around the central vein, while diffuse cytoplasmic staining is typical of β-catenin activation [1]. Nuclear β-catenin has low sensitivity for detecting β-catenin activation, while diffuse glutamine synthetase staining is more reliable [1, 3].
Although the use of diffuse glutamine synthetase as a surrogate for β-catenin activation has become standard practice to identify high risk tumors, the correlation of diffuse glutamine synthetase staining and β-catenin activation is not perfect. The reported correlation between diffuse glutamine synthetase staining and CTNNB1 mutations is 15–100% in hepatocellular carcinoma and 75–100% in atypical hepatocellular neoplasms [1,2,3,4, 15,16,17,18,19]. The reason for this discrepancy is not clear.
In this study, capture-based next-generation sequencing was performed in a series of atypical hepatocellular neoplasms and well-differentiated hepatocellular carcinoma with diffuse glutamine synthetase staining to examine the molecular basis of glutamine synthetase overexpression.
Materials and Methods
Cases
The study group comprised 13 atypical hepatocellular neoplasms and 14 well-differentiated hepatocellular carcinomas. Study cases were selected from the pathology files of the University of California, San Francisco and Southern California Permanente Medical Group. The study was approved by the institutional review boards. Hematoxylin and eosin stained sections and reticulin stains were reviewed in all cases, which were assigned to the categories enumerated below:
-
(1)
Atypical hepatocellular neoplasms (n = 13; all resections): These cases were divided into two groups:
-
(a)
Atypical hepatocellular neoplasm without atypia (n = 7): These cases showed morphologic features of hepatocellular adenoma with no cytoarchitectural atypia. These were considered atypical solely on the basis of presumed β-catenin activation due to diffuse glutamine synthetase staining.
-
(b)
Atypical hepatocellular neoplasm with cytoarchitectural atypia (n = 6): These tumors largely resembled hepatocellular adenoma but showed focal atypical morphological features such as small cell change, pseudoacinar architecture, thick cell plates and reticulin loss that were not considered sufficient for the diagnosis of hepatocellular carcinoma.
-
(a)
-
(2)
Well-differentiated hepatocellular carcinoma (n = 14; 13 resections, 1 biopsy): Tumors with obvious cytoarchitectural features of hepatocellular carcinoma such as thick cell plates, small cell change and reticulin loss. All cases were well-differentiated tumors according to the 2010 World Health Organization classification [20], and occurred in the setting of non-cirrhotic liver.
Immunohistochemistry
Immunohistochemical stains performed during the diagnostic work-up for the cases on whole sections from formalin-fixed paraffin-embedded tissue blocks were reviewed. The following antibodies were used: liver fatty acid binding protein, serum amyloid A, C-reactive protein, β-catenin and glutamine synthetase as previously described [2, 3].
Glutamine synthetase staining was considered diffuse if moderate to strong cytoplasmic staining was present in ≥ 50% of tumor cells, and was an inclusion criterion for all cases in this study. Diffuse glutamine synthetase staining was classified into one of the following two patterns: (a) diffuse homogeneous: moderate to strong cytoplasmic staining in more than 90% of lesional cells, without a map-like pattern, (b) diffuse heterogeneous: moderate to strong staining in 50–90% of lesional cells, without a map-like pattern. Nuclear β-catenin staining was interpreted as positive irrespective of number of positive cells, while membranous and/or cytoplasmic staining was considered negative. Positive nuclear β-catenin staining was further categorized, based on approximate number of positive tumor cells, as focal (< 5%), patchy (5–50%) and diffuse (>50%). Liver fatty acid binding protein staining was scored as present (normal result) or lost (abnormal result) based on cytoplasmic staining in the tumor cells; complete absence of staining in the tumor cells was required for liver fatty acid binding protein to be considered lost. Serum amyloid A and C-reactive protein were considered positive if moderate to strong cytoplasmic staining was seen in more than half of the tumor cells.
Capture-based next-generation sequencing and data analysis
Genomic DNA was extracted from matched normal and tumor formalin-fixed paraffin-embedded tissues from each case. Sequencing libraries were prepared from genomic DNA, target enrichment was performed by hybrid capture using custom baits designed to target all coding regions of 479 cancer-related genes, select introns from 47 genes, and the TERT promoter, with a total sequencing footprint of ~2.9 Mb (Supplementary Table S1). The baits also capture 2,000 unique sequences containing common single-nucleotide polymorphisms (SNPs) within regions devoid of constitutional copy number variations to assist in genome-wide copy number and allelic imbalance analysis. Sequencing was performed on an Illumina HiSeq 2500.
Duplicate sequencing reads were removed computationally to allow for accurate allele frequency determination and copy number calling. The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37), using the following software packages: BWA: 0.7.13, Samtools: 1.1 (using htslib 1.1), Picard tools: 1.97 (1504), GATK: Appistry v2015.1.1–3.4.46–0ga8e1d99, CNVkit: 0.7.2, Pindel: 0.2.5b8, SATK: Appistry v2015.1.1–1-gea45d62, Annovar: v2016 Feb01, Freebayes: 0.9.20 and Delly: 0.7.2 [21,22,23,24,25,26,27,28,29,30,31]. Somatic single-nucleotide variants, insertions/deletions, and rearrangements were visualized and verified using the Integrative Genomics Viewer (Broad Institute). Pathogenic and likely pathogenic somatic mutations were identified by first excluding all but non-synonymous, splice-site, and TERT promoter mutations. Next, we excluded variants that had >0.005 frequency in the Exome Sequencing Project (esp6500) (http://evs.gs.washington.edu/EVS/), Exome Aggregation Consortium (ExAc) [32] or the 1000Genomes database [33]. The remaining somatic variants were manually reviewed and annotated using knowledge present in the following databases: Catalogue of Somatic Mutations in Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic), cBioPortal for Cancer Genomics (www.cbioportal.org), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), and PubMed (https://www.ncbi.nlm.nih.gov/pubmed/). Genome-wide copy number analysis based on on-target and off-target reads was performed using CNVkit [27] and Nexus Copy Number (Biodiscovery, Hawthorne, CA). Focal copy number changes were defined as those encompassing less than 10 megabases. Large scale chromosomal changes were defined as those involving entire chromosomes or chromosome arms.
Results
Clinical features
Atypical hepatocellular neoplasms (4 males, 9 females, mean age 29 ± 14 years, age range 9–59 years) included 1 male patient with a history of anabolic steroid use, 2 female patients with a history of oral contraceptive use, and 1 male patient with Abernethy malformation (Supplementary Table S2). Two atypical hepatocellular neoplasms (9 and 10) were different nodules from the same patient, who had history of Ewing sarcoma. Patients with hepatocellular carcinoma (7 males, 7 females, mean age 36 ± 16 years, range 8–65 years) included 1 female patient with a history of oral contraceptive use. All atypical hepatocellular neoplasms and hepatocellular carcinomas occurred in non-cirrhotic liver.
Morphology and immunohistochemistry
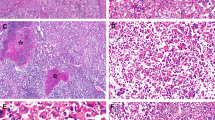
By definition, there were no cytoarchitectural abnormalities in 7 (54%) atypical hepatocellular neoplasms without atypia (Fig. 1). Cytoarchitectural atypia insufficient for a definite diagnosis of hepatocellular carcinoma was seen in 6 (46%) atypical hepatocellular neoplasms (Figs. 2 and 3). The focal atypical features included nuclear enlargement with prominent nucleoli (n = 2), crowding and small cell change (n = 3), focally thick cell plates (n = 3), pseudoacinar change (n = 2) and abnormal reticulin framework (n = 3). All hepatocellular carcinomas were well-differentiated and showed obvious cytoarchitectural atypia and abnormalities in reticulin framework (Figs. 4, 5). Inflammatory features in the form of positive serum amyloid A and/or C-reactive protein were seen in 8/10 (80%) atypical hepatocellular neoplasms; the results were not available in 3 cases. Loss of liver fatty acid binding protein was observed in 3 atypical hepatocellular neoplasms (2 from the same patient); all other atypical hepatocellular neoplasms showed intact liver fatty acid binding protein staining (Fig. 6, Supplementary Table S2).
Atypical hepatocellular neoplasm with inflammatory features showing no cytoarchitectural atypia (a, H&E stain ×200). Reticulin framework is partially disrupted, likely related to fat (b, Gomori reticulin stain, ×200). Serum amyloid A is positive (c, ×200) and glutamine synthetase shows diffuse homogeneous pattern (d, ×100) This tumor (atypical hepatocellular neoplasm #7) had an APC mutation without mutations involving the CTNNB1 gene
Atypical hepatocellular neoplasm with atypia characterized by nuclear enlargement, prominent nucleoli and focal crowding (a, H&E stain, ×200). Glutamine synthetase showed diffuse heterogeneous staining (b, ×100). This tumor (atypical hepatocellular neoplasm #11) showed S45 mutation in CTNNB1, which has been associated with diffuse heterogeneous glutamine synthetase staining (“starry sky” pattern)
Atypical hepatocellular neoplasm with atypia characterized by focally thick cell plates (a, H&E stain, ×200) and focal pseudoacinar architecture (b, H&E stain, ×200). Glutamine synthetase showed diffuse heterogeneous staining (c, ×200), and liver fatty acid binding protein was negative (d, ×200). This tumor (atypical hepatocellular neoplasm #9) showed HNF1A mutation, with no genomic alterations in the WNT signaling pathway
Well-differentiated hepatocellular carcinoma with thick cell plates, focal pseudoacinar architecture (a, H&E stain, ×200) and loss of reticulin network (b, Gomori reticulin stain, ×200). Glutamine synthetase showed diffuse heterogeneous staining (c, ×100) with peripheral accentuation (d, ×40). This tumor (hepatocellular carcinoma #2) showed mutations in AXIN1 and exon 7 of CTNNB1; the latter has been associated with peripheral glutamine synthetase staining
Genomic alterations in atypical hepatocellular neoplasms and well-differentiated hepatocellular carcinomas with diffuse glutamine synthetase staining. The top panel shows patient age, presence or absence of cytoarchitectural atypia, and immunohistochemical staining results for each case. The bottom panel provides a summary of all pathogenic and likely pathogenic mutations, as well as copy number alterations for each case
Diffuse glutamine synthetase staining was present in all 27 study cases. The staining pattern was diffuse homogeneous in 69% (9/13) and diffuse heterogeneous in 31% (4/13) of atypical hepatocellular neoplasms (Figs. 1–3, 6). In hepatocellular carcinomas, diffuse homogenous glutamine synthetase staining was seen in 79% (11/14) and diffuse heterogeneous in 21% (3/14) of cases (Figs. 4, 5, 6). Nuclear β-catenin staining was seen in 13/27 (48%) cases (Fig. 6). Nuclear β-catenin staining was seen in 5/13 (38%) atypical hepatocellular neoplasms, all of which had diffuse homogeneous glutamine synthetase staining. Of the 8 atypical hepatocellular neoplasms that lacked β-catenin nuclear staining, 4 each had diffuse homogeneous and diffuse heterogeneous glutamine synthetase staining. Nuclear β-catenin was seen in 8/14 (57%) hepatocellular carcinomas, all of which showed diffuse homogenous glutamine synthetase staining. Of the 6 hepatocellular carcinomas without nuclear β-catenin, 3 each showed diffuse homogeneous and diffuse heterogeneous glutamine synthetase staining.
Genomic alterations in atypical hepatocellular neoplasms and well-differentiated hepatocellular carcinomas with diffuse glutamine synthetase
The mean target sequencing coverage for atypical hepatocellular neoplasms was 562 ± 259 unique reads per target interval and for hepatocellular carcinomas was 457 ± 178 unique reads per target interval (Supplementary Table S3). Recurrent pathogenic and likely pathogenic alterations, as well as copy number changes are summarized in Fig. 6. Variant details, including genomic coordinates and mutant allele frequencies, are provided in Supplementary Table S4. Somatic variants of unknown significance are listed in Supplementary Table S5.
WNT pathway alterations
WNT pathway alterations were seen in 77% (10/13) of atypical hepatocellular neoplasms and 86% (12/14) of hepatocellular carcinoma (Table 1, 2, Fig. 6). CTNNB1 mutations or exon 3–4 deletions accounted for the majority of WNT pathway alterations in atypical hepatocellular neoplasms (9/10, 90%); one atypical hepatocellular neoplasm demonstrated a truncating mutation in APC with loss of heterozygosity. Three atypical hepatocellular neoplasms (two from the same patient), 1 with diffuse homogeneous and 2 with diffuse heterogeneous glutamine synthetase, as well as 1 hepatocellular carcinoma (diffuse heterogeneous glutamine synthetase) did not have WNT pathway alterations, but showed loss of liver fatty acid binding protein and HNF1A mutation (Figs. 3, 6). CTNNB1 mutations or exon 3–4 deletions were seen in 86% (12/14) of hepatocellular carcinomas. Mutations involving exon 7 of CTNNB1 were observed in 2 hepatocellular carcinoma cases, one of which also harbored an AXIN1 mutation; the other 10 cases involved exon 3 of CTNNB1. Of the 2 hepatocellular carcinomas without WNT pathway alterations, 1 had three mutations in HNF1A.
Other mutations and copy number changes
Aside from WNT pathway and HNF1A mutations, 54% (7/13) atypical hepatocellular neoplasms (4 without atypia, 3 with atypia) and 79% (11/14) of hepatocellular carcinomas had additional pathogenic alterations or chromosomal copy number alterations (Fig. 6). Three atypical hepatocellular neoplasms without atypia demonstrated additional truncating variants in tumor suppressor genes, with 1 each in TSC1, CREBBP, and DNMT3A. A fourth atypical hepatocellular neoplasm without atypia demonstrated loss of chromosome 8p and gain of chromosome 8q. Of the 6 atypical hepatocellular neoplasm with cytoarchitectural atypia, one case had a truncating TP53 mutation and loss of chromosome 17p, another case demonstrated gain of 8q and loss of distal 12q, and a third case demonstrated gain of chromosome 17. Of the 14 hepatocellular carcinoma cases, recurrent pathogenic mutations were seen in TERT promoter (3/14) and single cases demonstrated pathogenic variants in CDKN2A and MTOR. IDH1 mutations were observed in 2 hepatocellular carcinoma cases. Recurrent copy number alterations were also identified in the hepatocellular carcinoma cohort including gains in 1q (4 cases), 6 or 6p (2 cases), 7 (4 cases), 8q (2 cases), and 17q (1 case), as well as losses in 8p (2 cases), 12p (1 case), and 18q (1 case).
Correlation of glutamine synthetase staining pattern and mutational profile
Twenty out of twenty-seven cases had diffuse homogenous glutamine synthetase staining with 12 harboring CTNNB1 exon 3–4 in-frame deletions, 5 having CTNNB1 exon 3 mutations (one at I35/H36, two at S45, two at T41) and 1 with an APC truncating mutation; no WNT pathway alterations were seen in 2 cases. Of the seven cases with diffuse heterogeneous glutamine synthetase, two showed CTNNB1 exon 3 mutations (both at S45) and two had CTNNB1 exon 7 mutations (both at K335); no WNT pathway alterations were seen in 3 cases.
Comparison of atypical hepatocellular neoplasm and hepatocellular carcinoma
There was no significant difference in mutations in WNT signaling pathway, other mutations or copy number alterations in atypical hepatocellular neoplasm with and without atypia (Table 1). Similarly, WNT signaling alterations were similar in atypical hepatocellular neoplasm and hepatocellular carcinoma; TERT promoter mutations were seen only in hepatocellular carcinoma, and copy number changes were more common in hepatocellular carcinoma (Table 2).
Discussion
Activation of β-catenin is considered a high risk feature in well-differentiated hepatocellular tumors which do not fulfill the diagnostic criteria for hepatocellular carcinoma. Diffuse glutamine synthetase staining is commonly used as a marker of β-catenin in these atypical hepatocellular neoplasms. However, all cases with diffuse glutamine synthetase staining do not have CTNNB1 (β-catenin) mutations. Our study shows that alterations in the WNT signaling pathway were identified in 81% of tumors with diffuse glutamine synthetase (77% atypical hepatocellular neoplasms, 86% hepatocellular carcinomas). CTNNB1 exon 3 mutations/deletions accounted for the majority (70%) of cases, while CTNNB1 exon 7 and other WNT pathway components (APC, AXIN1) were involved in 11% of cases. If the analysis had focused only on CTNNB1 exon 3, these additional 11% of cases would have been missed. Diffuse glutamine synthetase staining has previously been reported with APC and AXIN mutations in hepatocellular carcinoma [34]. One hepatocellular carcinoma in our series showed both AXIN1 and exon 7 CTNNB1 mutation, and another hepatocellular carcinoma had an exon 7 CTNNB1 mutation as the only WNT pathway alteration. CTNNB1 exon 7 mutations do not typically lead to diffuse glutamine synthetase staining[4], and are thus thought to be associated with weak activation of β-catenin and lower risk of hepatocellular carcinoma; however, two hepatocellular carcinomas in our study had exon 7 mutations, and exon 7/8 mutations have been noted in 5% of hepatocellular carcinomas in one study [1].
The molecular basis for diffuse glutamine synthetase could not be determined in 19% of our cases (3 atypical hepatocellular neoplasms with atypia, 2 hepatocellular carcinomas). Although perfect correlation between diffuse glutamine synthetase and CTNNB1 has been described in some studies [1], this lack of correlation has been noted in other studies in both atypical hepatocellular neoplasms and hepatocellular carcinomas [3, 15,16,17, 19]. Diffuse glutamine synthetase staining has been observed in 60–70% of hepatocellular carcinomas in the literature, while β-catenin mutations occur in only 15–35% of hepatocellular carcinomas [19]. In a prior study, we found a 75% correlation between diffuse glutamine synthetase staining and CTNNB1 mutations in atypical hepatocellular neoplasms [3]; however, other WNT signaling pathway genes were not examined and some cases with CTNNB1 exon 3 large deletions may have been missed as Sanger sequencing from paraffin-embedded tissue was used in that study. It is possible that the WNT signaling pathway is activated in these cases through other mechanisms that were not examined by our assay. Diffuse glutamine synthetase staining has also been postulated to occur through other mechanisms such as alterations in blood flow, oxygenation, nutritional status, cholestasis, and fibrosis/cirrhosis [35,36,37]. Expansion of glutamine synthetase staining (map-like pattern) occurs in focal nodular hyperplasia without mutations in CTNNB1 or other WNT signaling genes [14, 38]. Occasional cirrhotic nodules can show diffuse glutamine synthetase staining without clear evidence of β-catenin activation (personal observation). It is not known if any of these mechanisms can explain the diffuse glutamine synthetase staining in atypical hepatocellular neoplasm cases.
It is intriguing that 3 atypical hepatocellular neoplasms (including 2 from the same patient) and 1 hepatocellular carcinoma in our series showed diffuse glutamine synthetase (heterogeneous in 3, homogeneous in 1) without genetic alterations in the WNT signaling pathway, but harbored HNF1A mutations. The presence of simultaneous CTNNB1 and HNF1A mutations in the same tumor has been postulated to explain this phenomenon [3, 39]. However, none of the 4 tumors in our study with HNF1A mutations and diffuse glutamine synthetase had CTNNB1 mutations, and the mechanism of diffuse glutamine synthetase in these cases remains elusive. Loss of liver fatty acid-binding protein has been reported in hepatocellular carcinoma but neither mutation analysis nor staining glutamine synthetase was examined in these cases [40].
Diffuse homogenous glutamine synthetase staining (≥90% of tumor cells) is thought to result from strong β-catenin activation, often due to large deletions, point mutations in the β-TrCP binding domain (D32-S37) and T41 mutation in exon 3 of the CTNNB1 gene [4]. Diffuse heterogeneous glutamine synthetase staining (≥50% but <90% of tumor cells) indicates relatively weaker β-catenin activation, often as a result of S45 mutations in exon 3 CTNNB1, while exon 7 and 8 mutations lead to even weaker activation with patchy and not diffuse glutamine synthetase staining [3, 4]. We have earlier pointed out that the correlation of glutamine synthetase patterns and CTNNB1 mutations are not perfect [3, 41]. Our results show that exon 3 deletions, D32-S37, and T41 mutations lead to diffuse homogeneous pattern, but this can also be seen with S45 mutation, while diffuse heterogeneous pattern can be observed with exon 7 mutations. The presence of CTNNB1 exon 7 mutation in 2 hepatocellular carcinomas and their association with diffuse heterogeneous glutamine synthetase suggests that these mutations may not be benign as earlier suggested [3, 4], even though the risk may be lower compared to CTNNB1 exon 3 mutation. This is further supported by CTNNB1 exon7/8 mutations occurring in 5% of hepatocellular carcinomas in a large series [4, 41].
In this study, atypical hepatocellular neoplasms were divided into two categories based on presence or absence of cytoarchitectural features. The pathogenic mutations and cytogenetic abnormalities in these categories were similar. We also observed overlap of mutations and copy number changes in atypical hepatocellular neoplasms and hepatocellular carcinomas, emphasizing the close relationship of these tumors. Mutations such as TP53 and TSC1 that have been well described in hepatocellular carcinoma were observed in atypical hepatocellular neoplasms [42]. Copy number changes in atypical hepatocellular neoplasms included 8q gain, 8p loss, chromosome 17 gain, 17p loss, and 12p loss. These changes, especially those involving chromosome 8, are common in hepatocellular carcinoma, including well-differentiated cases [2, 8, 9, 43, 44]. There were differences in atypical hepatocellular neoplasms and hepatocellular carcinomas as well indicating a more progressed nature of the latter. These changes included more frequent copy number alterations in well-differentiated hepatocellular carcinomas (64 vs. 31%), and mutations involving TERT promoter and IDH1 that were observed exclusively in hepatocellular carcinoma. TERT promoter mutation has been observed in 17% of borderline lesions compared to 54% of hepatocellular carcinomas [45]. The differences in definitions of borderline lesions in this study and atypical hepatocellular neoplasms in our study is likely responsible for the reported 17% TERT promoter rate in borderline lesions compared to none of atypical hepatocellular neoplasm cases in our series. IDH1 mutation is characteristic of intrahepatic cholangiocarcinoma [46, 47], but has been reported in hepatocellular carcinoma [45]. The comparison of atypical hepatocellular neoplasms and hepatocellular carcinomas in this study is based on a limited number of cases, but the results are somewhat similar to our earlier study using fluorescence in situ hybridization as well as the study from the French group [2, 45]. Similar comparison in larger studies is necessary to corroborate these findings.
In conclusion, this study shows that CTNNB1 exon 3 deletion/mutation accounts for the diffuse glutamine synthetase staining in well-differentiated hepatocellular neoplasms in ~70% of cases, while mutation in other exons of CTNNB1 and other WNT signaling alterations is observed in approximately 11% of cases. If sequencing assays are used to evaluate for β-catenin activation in tumors with indeterminate glutamine synthetase staining, exons 7/8 and other WNT signaling components like APC and AXIN should be examined in addition to exon 3 of CTNNB1. The mechanism of diffuse glutamine synthetase remained elusive in nearly one-fifth of atypical hepatocellular neoplasm/ hepatocellular carcinoma; it is not clear whether diffuse glutamine synthetase is a high risk feature in such cases. The similarity of atypical hepatocellular neoplasm with and without atypia in terms of WNT signaling activation, other pathogenic mutations and cytogenetic changes supports the proposal that β-catenin-activated tumors should not be designated as hepatocellular adenoma, but placed in a new ‘atypical’ category irrespective of morphology. Additional pathogenic alterations like mutations in TERT promoter/IDH1/CDKN2A and cytogenetic abnormalities in this setting strongly favor the diagnosis of hepatocellular carcinoma. Clinical assays combining WNT pathway mutations (CTNNB1, APC, AXIN), TERT promoter mutation and cytogenetic assays for selected gains/losses can be helpful in diagnosis and risk stratification of hepatocellular tumors in challenging settings when morphology and glutamine synthetase immunohistochemistry do not provide a definite diagnosis.
References
Bioulac-Sage P, Rebouissou S, Thomas C, et al. Hepatocellular adenoma subtype classification using molecular markers and immunohistochemistry. Hepatology. 2007;46:740–8.
Evason KJ, Grenert JP, Ferrell LD, et al. Atypical hepatocellular adenoma-like neoplasms with beta-catenin activation show cytogenetic alterations similar to well-differentiated hepatocellular carcinomas. Hum Pathol. 2013;44:750–8.
Hale G, Liu X, Hu J, et al. Correlation of exon 3 beta-catenin mutations with glutamine synthetase staining patterns in hepatocellular adenoma and hepatocellular carcinoma. Mod Pathol. 2016;29:1370–80.
Rebouissou S, Franconi A, Calderaro J, et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology. 2016;64:2047–61.
Zucman-Rossi J, Jeannot E, Nhieu JT, et al. Genotype-phenotype correlation in hepatocellular adenoma: new classification and relationship with HCC. Hepatology. 2006;43:515–24.
Bioulac-Sage P, Balabaud C, Wanless I. Focal nodular hyperplasia and hepatocellular adenoma. In: Bosman FT, Carneiro F, Hruban RH and Theise ND, editors. WHO classification of tumors of the digestive system. 4th ed. Lyon: IARC Press, 2010. p. 198-204.
Farges O, Ferreira N, Dokmak S, et al. Changing trends in malignant transformation of hepatocellular adenoma. Gut. 2011;60:85–89.
Kakar S, Chen X, Ho C, et al. Chromosomal abnormalities determined by comparative genomic hybridization are helpful in the diagnosis of atypical hepatocellular neoplasms. Histopathology. 2009;55:197–205.
Pilati C, Letouze E, Nault JC, et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell. 2014;25:428–41.
Balabaud C, Bioulac-Sage P, Ferrell L, et al. Well-differentiated hepatocellular neoplasm of uncertain malignant potential. Hum Pathol. 2015;46:634–5.
Bedossa P, Burt AD, Brunt EM, et al. Well-differentiated hepatocellular neoplasm of uncertain malignant potential: proposal for a new diagnostic category. Hum Pathol. 2014;45:658–60.
Kakar S, Evason KJ, Ferrell LD. Well-differentiated hepatocellular neoplasm of uncertain malignant potential: proposal for a new diagnostic category–reply. Hum Pathol. 2014;45:660–1.
European Association for the Study of the Liver. EASL clinical practice guidelines on the management of benign liver tumours. J Hepatol. 2016;65:386–98.
Joseph NM, Ferrell LD, Jain D, et al. Diagnostic utility and limitations of glutamine synthetase and serum amyloid-associated protein immunohistochemistry in the distinction of focal nodular hyperplasia and inflammatory hepatocellular adenoma. Mod Pathol. 2014;27:62–72.
Audard V, Grimber G, Elie C, et al. Cholestasis is a marker for hepatocellular carcinomas displaying beta-catenin mutations. J Pathol. 2007;212:345–52.
Austinat M, Dunsch R, Wittekind C, et al. Correlation between beta-catenin mutations and expression of Wnt-signaling target genes in hepatocellular carcinoma. Mol Cancer. 2008;7:21.
Dal Bello B, Rosa L, Campanini N, et al. Glutamine synthetase immunostaining correlates with pathologic features of hepatocellular carcinoma and better survival after radiofrequency thermal ablation. Clin Cancer Res. 2010;16:2157–66.
Kakar S, Grenert JP, Paradis V, et al. Hepatocellular carcinoma arising in adenoma: similar immunohistochemical and cytogenetic features in adenoma and hepatocellular carcinoma portions of the tumor. Mod Pathol. 2014;27:1499–509.
Prange W, Breuhahn K, Fischer F, et al. Beta-catenin accumulation in the progression of human hepatocarcinogenesis correlates with loss of E-cadherin and accumulation ofp53, but not with expression of conventional WNT-1 target genes. J Pathol. 2003;201:250–9.
Thiese ND, Curado MP, Franceshchi S, et al. Hepatocellular carcinoma. In: Bosman FT, Carneiro F, Hruban RH and Theise ND, editors. WHO classification of tumors of the digestive system. 4th ed. Lyon: IARC Press, 2010. p. 198-204.
DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. eprint arXiv:1207.3907 [q-bio.GN].
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Li H, Handsaker B, Wysoker A, et al. The Sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Rausch T, Zichner T, Schlattl A, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i1339.
Talevich E, Shain AH, Botton T, et al. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12:e1004873.
Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10. 1–11.10.33.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66.
Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Genomes Project C, Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73.
Zucman-Rossi J, Benhamouche S, Godard C, et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–80.
Arnason T, Fleming KE, Wanless IR. Peritumoral hyperplasia of the liver: a response to portal vein invasion by hypervascular neoplasms. Histopathology. 2013;62:458–64.
Paxian M, Rensing H, Geckeis K, et al. Perflubron emulsion in prolonged hemorrhagic shock: influence on hepatocellular energy metabolism and oxygen-dependent gene expression. Anesthesiology. 2003;98:1391–9.
Ueberham E, Arendt E, Starke M, et al. Reduction and expansion of the glutamine synthetase expressing zone in livers from tetracycline controlled TGF-beta1 transgenic mice and multiple starved mice. J Hepatol. 2004;41:75–81.
Bioulac-Sage P, Laumonier H, Rullier A, et al. Over-expression of glutamine synthetase in focal nodular hyperplasia: a novel easy diagnostic tool in surgical pathology. Liver Int. 2009;29:459–65.
Shafizadeh N, Genrich G, Ferrell L, et al. Hepatocellular adenomas in a large community population, 2000 to 2010: reclassification per current World Health Organization classification and results of long-term follow-up. Hum Pathol. 2014;45:976–83.
Cho SJ, Ferrell LD, Gill RM. Expression of liver fatty acid binding protein in hepatocellular carcinoma. Hum Pathol. 2016;50:135–9.
Kakar S, Ferrell LD. Glutamine synthetase staining and CTTNB1 mutation in hepatocellular adenomas. Hepatology. 2017;66:2092–3.
Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340–9.
Balsara BR, Pei J, De Rienzo A, et al. Human hepatocellular carcinoma is characterized by a highly consistent pattern of genomic imbalances, including frequent loss of 16q23.1-24.1. Genes Chromosomes Cancer. 2001;30:245–53.
Wilkens L, Bredt M, Flemming P, et al. Differentiation of liver cell adenomas from well-differentiated hepatocellular carcinomas by comparative genomic hybridization. J Pathol. 2001;193:476–82.
Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–41.
Farshidfar F, Zheng S, Gingras MC, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep. 2017;19:2878–80.
Kipp BR, Voss JS, Kerr SE, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol. 2012;43:1552–8.
Funding
This study was funded by the University of California San Francisco Department of Pathology Research Endowment awards to NMJ and SK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Joseph, N.M., Umetsu, S.E., Shafizadeh, N. et al. Genomic profiling of well-differentiated hepatocellular neoplasms with diffuse glutamine synthetase staining reveals similar genetics across the adenoma to carcinoma spectrum. Mod Pathol 32, 1627–1636 (2019). https://doi.org/10.1038/s41379-019-0282-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0282-0