Abstract
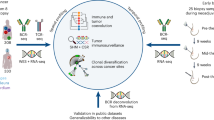
Complete elimination of B-cell acute lymphoblastic leukemia (B-ALL) by a risk-adapted primary treatment approach remains a clinical key objective, which fails in up to a third of patients. Recent evidence has implicated subpopulations of B-ALL cells with stem-like features in disease persistence. We hypothesized that microRNA-126, a core regulator of hematopoietic and leukemic stem cells, may resolve intratumor heterogeneity in B-ALL and uncover therapy-resistant subpopulations. We exploited patient-derived xenograft (PDX) models with B-ALL cells transduced with a miR-126 reporter allowing the prospective isolation of miR-126(high) cells for their functional and transcriptional characterization. Discrete miR-126(high) populations, often characterized by MIR126 locus demethylation, were identified in 8/9 PDX models and showed increased repopulation potential, in vivo chemotherapy resistance and hallmarks of quiescence, inflammation and stress-response pathway activation. Cells with a miR-126(high) transcriptional profile were identified as distinct disease subpopulations by single-cell RNA sequencing in diagnosis samples from adult and pediatric B-ALL. Expression of miR-126 and locus methylation were tested in several pediatric and adult B-ALL cohorts, which received standardized treatment. High microRNA-126 levels and locus demethylation at diagnosis associate with suboptimal response to induction chemotherapy (MRD > 0.05% at day +33 or MRD+ at day +78).

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
ENA code for Bisulphite targeted sequencing is PRJEB63552. ENA code for Whole exome sequencing is PRJEB63520. Bulk RNA sequencing data can be found under the GEO superseries code GSE236142.
Code availability
All codes are available at the following git repository http://www.bioinfotiget.it/gitlab/custom/casertanucera_leukemia2023.
References
Lanza F, Maffini E, Saraceni F, Massari E, Rondoni M, Daghia G, et al. New monoclonal antibodies and tyrosine kinase inhibitors in B-cell acute lymphoblastic leukemia. Minerva Med. 2020;111:478–90. https://pubmed.ncbi.nlm.nih.gov/32955830/.
Rowe JM, Buck G, Burnett AK, Chopra R, Wiernik PH, Richards SM, et al. Induction therapy for adults with acute lymphoblastic leukemia: results of more than 1500 patients from the international ALL trial: MRC UKALL XII/ECOG E2993. Blood. 2005;106:3760–7.
Liew E, Atenafu EG, Schimmer AD, Yee KWL, Schuh AC, Minden MD, et al. Outcomes of adult patients with relapsed acute lymphoblastic leukemia following frontline treatment with a pediatric regimen. Leuk Res. 2012;36:1517–20. http://www.sciencedirect.com/science/article/pii/S0145212612003529.
Kim IS. Minimal residual disease in acute lymphoblastic leukemia: technical aspects and implications for clinical interpretation. Blood Res. 2020;55:S19–26. https://www.bloodresearch.or.kr/journal/view.html?doi=10.5045/br.2020.S004.
Sikich N, Soulodre C, Sadasook N, Sleeman A, Verhey J. Citation Health Quality Ontario. Minimal residual disease evaluation in childhood acute lymphoblastic leukemia: a clinical evidence review. Ont Health Technol Assess Ser. Ont Health Technol Assess Ser. 2016;16:1–52.
van Dongen JJM, van der Velden VHJ, Brüggemann M, Orfao A. Minimal residual disease diagnostics in acute lymphoblastic leukemia: Need for sensitive, fast, and standardized technologies. Blood. 2015;125:3996–4009. https://pubmed.ncbi.nlm.nih.gov/25999452/.
Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–91. https://doi.org/10.1016/j.stem.2014.02.006.
Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–80.
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604 https://pubmed.ncbi.nlm.nih.gov/25790293.
Dobson SM, García-Prat L, Vanner RJ, Wintersinger J, Waanders E, Gu Z, et al. Relapse-fated latent diagnosis subclones in acute B lineage leukemia are drug tolerant and possess distinct metabolic programs. Cancer Discov. 2020;10:568–87.
Waanders E, Gu Z, Dobson SM, Antić Ž, Crawford JC, Ma X. et al. Mutational landscape and patterns of clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Blood Cancer Discov. 2020;1:96–111.
Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science (1979). 2017;357:eaal2380.
Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. http://www.ncbi.nlm.nih.gov/pubmed/23142521.
Lechman ER, Gentner B, Ng SWK, Schoof EM, van Galen P, Kennedy JA. et al. MiR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells [Cancer Cell 29, (2016) 214-228; February 8]. Cancer Cell. 2016;29:602–6.
Gentner B, Visigalli I, Hiramatsu H, Lechman E, Ungari S, Giustacchini A, et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med. 2010;2:58ra84–58ra84. http://www.ncbi.nlm.nih.gov/pubmed/21084719.
Li Z, Chen P, Su R, Li Y, Hu C, Wang Y, et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood. 2015;126:2005–15.
Zhang B, Nguyen LXT, Li L, Zhao D, Kumar B, Wu H, et al. Bone marrow niche trafficking of miR-126 controls the self-renewal of leukemia stem cells in chronic myelogenous leukemia. Nat Med. 2018;24:450–62. https://doi.org/10.1038/nm.4499.
Nucera S, Giustacchini A, Boccalatte F, Calabria A, Fanciullo C, Plati T, et al. miRNA-126 orchestrates an oncogenic program in B cell precursor acute lymphoblastic leukemia. Cancer Cell. 2016;29:905–21.
Lechman ER, Gentner B, Ng SWK, Schoof EM, van Galen P, Kennedy JA, et al. MiR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29:214–28.
de Leeuw DC, Denkers F, Olthof MC, Rutten AP, Pouwels W, Jan Schuurhuis G, et al. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014;74:2094–105. http://www.ncbi.nlm.nih.gov/pubmed/24477595.
Harnprasopwata R, Hab D, Toyoshimaa T, Lodishc H, Tojoa A. Alteration of processing induced by a single nucleotide polymorphism in pri-miR-126. Biochem Biophys Res Commun. 2010;399:117–22.
Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505.
Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, et al. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell. 2016;167:219–232.e14. https://www.sciencedirect.com/science/article/pii/S0092867416312363?via%3Dihub.
Lei H, Quelle FW. FOXO transcription factors enforce cell cycle checkpoints and promote survival of hematopoietic cells after DNA damage. Mol Cancer Res. 2009;7:1294–303. http://mcr.aacrjournals.org/.
Laurenti E, Doulatov S, Zandi S, Plumb I, Chen J, April C, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol. 2013;14:756–63.
McClellan JS, Dove C, Gentles AJ, Ryan CE, Majeti R. Reprogramming of primary human Philadelphia chromosome-positive B cell acute lymphoblastic leukemia cells into nonleukemic macrophages. Proc Natl Acad Sci USA. 2015;112:4074–9.
Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–41. https://pubmed.ncbi.nlm.nih.gov/21390130/.
Schwartzman O, Savino AM, Gombert M, Palmi C, Cario G, Schrappe M, et al. Suppressors and activators of JAK-STAT signaling at diagnosis and relapse of acute lymphoblastic leukemia in Down syndrome. Proc Natl Acad Sci USA. 2017;114:E4030–9. https://pubmed.ncbi.nlm.nih.gov/28461505/.
Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2016;113:11306–11. https://pubmed.ncbi.nlm.nih.gov/27655895/.
Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun. 2014;5. https://pubmed.ncbi.nlm.nih.gov/24662245/.
Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45:290–4. https://pubmed.ncbi.nlm.nih.gov/23377183/.
Li B, Brady SW, Ma X, Shen S, Zhang Y, Li Y, et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood. 2020;135:41–55. https://pubmed.ncbi.nlm.nih.gov/31697823/.
Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–7.
Ebinger S, Özdemir EZ, Ziegenhain C, Tiedt S, Castro Alves C, Grunert M, et al. Characterization of rare, dormant, and therapy-resistant cells in acute lymphoblastic leukemia. Cancer Cell. 2016;30:849–62.
Turati VA, Guerra-Assunção JA, Potter NE, Gupta R, Ecker S, Daneviciute A, et al. Chemotherapy induces canalization of cell state in childhood B-cell precursor acute lymphoblastic leukemia. Nat Cancer. 2021;2:835–52. https://www.nature.com/articles/s43018-021-00219-3.
Hu Y, Liu Y, Pelletier S, Buchdunger E, Warmuth M, Fabbro D, et al. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat Genet. 2004;36:453–61.
Acknowledgements
We thank all members from the Gentner lab for help with experiments, discussion and useful insight; all Fractal and Alembic facility personnel for cell sorting and imaging; the center for Omics sciences (COSR) for advice and assistance with genomic sequencing; the OSR biobank for sample collection and storage; the Hematology department for fruitful discussions. We acknowledge Cristina Bugarin for sorting of pediatric patient samples. We thank Matteo Massidda from CRS, Cagliari, Italy, for help with bioinformatic analysis.
Funding
This research was supported by grants to B.G. from the Italian Association for Cancer Research (AIRC-IG 2018 Id.22143), a John Goldman Clinical Research Fellowship from the European Hematology Association (EHA 2014) and from the Telethon Foundation (TIGET 2016 core grant no. C1). Research was also supported by a grant to A.B. from the Italian Association for Cancer Research (AIRC-IG 2017 Id.20564).
Author information
Authors and Affiliations
Contributions
C.C. planned and performed experiments, analyzed and interpreted data and prepared the manuscript; S.N. planned and performed experiments, analyzed and interpreted data and collected clinical data. M.Ba. developed and performed the bioinformatics analyses; G.F. and M.D.A. performed sequencing and analyzed data; M.M.N. performed experiments and helped in setting up scRNAseq; F.P. assisted with patient sample identification and collection; G.D. E.Z. and P.C. provided technical assistance and experimental expertise; A.L., I.M. provided supervision; D.S. and M.G.V. provided statistical analysis; F.C., O.S., A.R., A.B. provided supervision and access to patient cells. G.C. provided intellectual input, funding and research infrastructure; B.G. provided funding, designed and coordinated the research, interpreted the data, supervised research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Caserta, C., Nucera, S., Barcella, M. et al. miR-126 identifies a quiescent and chemo-resistant human B-ALL cell subset that correlates with minimal residual disease. Leukemia 37, 1994–2005 (2023). https://doi.org/10.1038/s41375-023-02009-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-02009-5