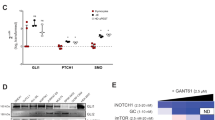
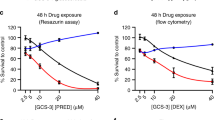
Abstract
Despite decades of clinical use, mechanisms of glucocorticoid resistance are poorly understood. We treated primary murine T lineage acute lymphoblastic leukemias (T-ALLs) with the glucocorticoid dexamethasone (DEX) alone and in combination with the pan-PI3 kinase inhibitor GDC-0941 and observed a robust response to DEX that was modestly enhanced by GDC-0941. Continuous in vivo treatment invariably resulted in outgrowth of drug-resistant clones, ~30% of which showed markedly reduced glucocorticoid receptor (GR) protein expression. A similar proportion of relapsed human T-ALLs also exhibited low GR protein levels. De novo or preexisting mutations in the gene encoding GR (Nr3c1) occurred in relapsed clones derived from multiple independent parental leukemias. CRISPR/Cas9 gene editing confirmed that loss of GR expression confers DEX resistance. Exposing drug-sensitive T-ALLs to DEX in vivo altered transcript levels of multiple genes, and this response was attenuated in relapsed T-ALLs. These data implicate reduced GR protein expression as a frequent cause of glucocorticoid resistance in T-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gao J, Liu WJ. Prognostic value of the response to prednisone for children with acute lymphoblastic leukemia: a meta-analysis. Eur Rev Med Pharmacol Sci. 2018;22:7858–66.
Pui CH, Dahl GV, Rivera G, Murphy SB, Costlow ME. The relationship of blast cell glucocorticoid receptor levels to response to single-agent steroid trial and remission response in children with acute lymphoblastic leukemia. Leuk Res. 1984;8:579–85.
Pui CH, Ochs J, Kalwinsky DK, Costlow ME. Impact of treatment efficacy on the prognostic value of glucocorticoid receptor levels in childhood acute lymphoblastic leukemia. Leuk Res. 1984;8:345–50.
Pui CH, Costlow ME. Sequential studies of lymphoblast glucocorticoid receptor levels at diagnosis and relapse in childhood leukemia: an update. Leuk Res. 1986;10:227–9.
Lauten M, Cario G, Asgedom G, Welte K, Schrappe M. Protein expression of the glucocorticoid receptor in childhood acute lymphoblastic leukemia. Haematologica. 2003;88:1253–8.
Ashraf J, Thompson EB. Identification of the activation-labile gene: a single point mutation in the human glucocorticoid receptor presents as two distinct receptor phenotypes. Mol Endocrinol. 1993;7:631–42.
Powers JH, Hillmann AG, Tang DC, Harmon JM. Cloning and expression of mutant glucocorticoid receptors from glucocorticoid-sensitive and -resistant human leukemic cells. Cancer Res. 1993;53:4059–65.
Strasser-Wozak EM, Hattmannstorfer R, Hala M, Hartmann BL, Fiegl M, Geley S, et al. Splice site mutation in the glucocorticoid receptor gene causes resistance to glucocorticoid-induced apoptosis in a human acute leukemic cell line. Cancer Res. 1995;55:348–53.
Geley S, Hartmann BL, Hala M, Strasser-Wozak EM, Kapelari K, Kofler R. Resistance to glucocorticoid-induced apoptosis in human T-cell acute lymphoblastic leukemia CEM-C1 cells is due to insufficient glucocorticoid receptor expression. Cancer Res. 1996;56:5033–8.
Hala M, Hartmann BL, Bock G, Geley S, Kofler R. Glucocorticoid-receptor-gene defects and resistance to glucocorticoid-induced apoptosis in human leukemic cell lines. Int J Cancer. 1996;68:663–8.
Riml S, Schmidt S, Ausserlechner MJ, Geley S, Kofler R. Glucocorticoid receptor heterozygosity combined with lack of receptor auto-induction causes glucocorticoid resistance in Jurkat acute lymphoblastic leukemia cells. Cell Death Differ. 2004;11(Suppl 1):S65–72.
Schmidt S, Irving JA, Minto L, Matheson E, Nicholson L, Ploner A, et al. Glucocorticoid resistance in two key models of acute lymphoblastic leukemia occurs at the level of the glucocorticoid receptor. FASEB J. 2006;20:2600–2.
Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–80.
Hogan LE, Meyer JA, Yang J, Wang J, Wong N, Yang W, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118:5218–26.
Kunz JB, Rausch T, Bandapalli OR, Eilers J, Pechanska P, Schuessele S, et al. Pediatric T-cell lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica. 2015;100:1442–50.
Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2016;113:11306–11.
Spinella JF, Richer C, Cassart P, Ouimet M, Healy J, Sinnett D. Mutational dynamics of early and late relapsed childhood ALL: rapid clonal expansion and long-term dormancy. Blood Adv. 2018;2:177–88.
Kuster L, Grausenburger R, Fuka G, Kaindl U, Krapf G, Inthal A, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood. 2011;117:2658–67.
Bokemeyer A, Eckert C, Meyr F, Koerner G, von Stackelberg A, Ullmann R, et al. Copy number genome alterations are associated with treatment response and outcome in relapsed childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Haematologica. 2014;99:706–14.
Grausenburger R, Bastelberger S, Eckert C, Kauer M, Stanulla M, Frech C, et al. Genetic alterations in glucocorticoid signaling pathway components are associated with adverse prognosis in children with relapsed ETV6/RUNX1-positive acute lymphoblastic leukemia. Leuk Lymphoma. 2016;57:1163–73.
Irving JA, Enshaei A, Parker CA, Sutton R, Kuiper RP, Erhorn A, et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2016;128:911–22.
Lauchle JO, Kim D, Le DT, Akagi K, Crone M, Krisman K, et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461:411–4.
Dail M, Li Q, McDaniel A, Wong J, Akagi K, Huang B, et al. Mutant Ikzf1, KrasG12D, and Notch1 cooperate in T lineage leukemogenesis and modulate responses to targeted agents. Proc Natl Acad Sci USA. 2010;107:5106–11.
Dail M, Wong J, Lawrence J, O'Connor D, Nakitandwe J, Chen SC, et al. Loss of oncogenic Notch1 with resistance to a PI3K inhibitor in T-cell leukaemia. Nature. 2014;513:512–6.
Burgess MR, Hwang E, Mroue R, Bielski CM, Wandler AM, Huang BJ, et al. KRAS allelic imbalance enhances fitness and modulates MAP kinase dependence in cancer. Cell. 2017;168:817–29. e15.
Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, et al. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell. 2013;24:766–76.
Wolff L, Koller R, Hu X, Anver MR. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J Virol. 2003;77:4965–71.
Huang BJ, Wandler AM, Meyer LK, Dail M, Daemen A, Sampath D, et al. Convergent genetic aberrations in murine and human T lineage acute lymphoblastic leukemias. PLoS Genet. 2019;15:e1008168.
Norman MR, Thompson EB. Characterization of a glucocorticoid-sensitive human lymphoid cell line. Cancer Res. 1977;37:3785–91.
Mansour MR, Duke V, Foroni L, Patel B, Allen CG, Ancliff PJ, et al. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2007;13:6964–9.
Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–71.
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604.
Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sanchez-Martin M, Madubata CJ, et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature. 2018;553:511–4.
Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–71.
Schwartz CL, Thompson EB, Gelber RD, Young ML, Chilton D, Cohen HJ, et al. Improved response with higher corticosteroid dose in children with acute lymphoblastic leukemia. J Clin Oncol. 2001;19:1040–6.
Haarman EG, Kaspers GJ, Pieters R, Rottier MM, Veerman AJ. Glucocorticoid receptor alpha, beta and gamma expression vs in vitro glucocorticod resistance in childhood leukemia. Leukemia. 2004;18:530–7.
Tissing WJ, Meijerink JP, Brinkhof B, Broekhuis MJ, Menezes RX, den Boer ML, et al. Glucocorticoid-induced glucocorticoid-receptor expression and promoter usage is not linked to glucocorticoid resistance in childhood ALL. Blood. 2006;108:1045–9.
Gruber G, Carlet M, Turtscher E, Meister B, Irving JA, Ploner C, et al. Levels of glucocorticoid receptor and its ligand determine sensitivity and kinetics of glucocorticoid-induced leukemia apoptosis. Leukemia. 2009;23:820–3.
Xiao H, Ding Y, Gao Y, Wang LM, Wang H, Ding L, et al. Haploinsufficiency of NR3C1 drives glucocorticoid resistance in adult acute lymphoblastic leukemia cells by down-regulating the mitochondrial apoptosis axis, and is sensitive to Bcl-2 blockage. Cancer Cell Int. 2019;19:218.
Li B, Brady SW, Ma X, Shen S, Zhang Y, Li Y, et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood. 2020;135:41–55.
Paugh SW, Bonten EJ, Savic D, Ramsey LB, Thierfelder WE, Gurung P, et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat Genet. 2015;47:607–14.
Li J, Troche C, Swaroop A, Kulis M, Oyer J, Will C, et al. A gain of function mutation in the NSD2 histone methyltransferase drives glucocorticoid resistance of acute lymphoblastic leukemia. Blood. 2018;132(Suppl 1):653.
Li B, Li H, Bai Y, Kirschner-Schwabe R, Yang JJ, Chen Y, et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med. 2015;21:563–71.
Jamieson CA, Yamamoto KR. Crosstalk pathway for inhibition of glucocorticoid-induced apoptosis by T cell receptor signaling. Proc Natl Acad Sci USA. 2000;97:7319–24.
Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80.
Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun. 2016;7:10690.
Raha D, Wilson TR, Peng J, Peterson D, Yue P, Evangelista M, et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res. 2014;74:3579–90.
Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–9.
Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–50.
Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–70.
Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24.
Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44.
Delgado-Martin C, Meyer LK, Huang BJ, Shimano KA, Zinter MS, Nguyen JV, et al. JAK/STAT pathway inhibition overcomes IL7-induced glucocorticoid resistance in a subset of human T-cell acute lymphoblastic leukemias. Leukemia. 2017;31:2568–76.
Meyer LK, Huang BJ, Delgado-Martin C, Roy RP, Hechmer A, Wandler AM, et al. Glucocorticoids paradoxically facilitate steroid resistance in T-cell acute lymphoblastic leukemias and thymocytes. J Clin Investig. 2020;130:863–76.
Mansour MR, He S, Li Z, Lobbardi R, Abraham BJ, Hug C, et al. JDP2: An oncogenic bZIP transcription factor in T cell acute lymphoblastic leukemia. J Exp Med. 2018;215:1929–45.
Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7.
Jing D, Huang Y, Liu X, Sia KCS, Zhang JC, Tai X, et al. Lymphocyte-specific chromatin accessibility pre-determines glucocorticoid resistance in acute lymphoblastic leukemia. Cancer Cell. 2018;34:906–21. e8.
Burgess MR, Hwang E, Firestone AJ, Huang T, Xu J, Zuber J, et al. Preclinical efficacy of MEK inhibition in Nras-mutant AML. Blood. 2014;124:3947–55.
Uren AG, Mikkers H, Kool J, van der Weyden L, Lund AH, Wilson CH, et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc. 2009;4:789–98.
Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168.
Acknowledgements
We thank members of the Integrated Genomics Operation in the Marie-Josee and Henry R. Kravis Center for Molecular Oncology at the Memorial Sloan-Kettering Cancer Center, and the Genome Sequencing Lab at St. Jude Children’s Research Hospital for assistance with sequencing; the UCSF Biorepository and Tissue Biomarker Technology Core and the DF/HCC Research Pathology Cores at Harvard Medical School for performing immunohistochemistry analysis. This study was supported in part by the UCSF Helen Diller Family Comprehensive Cancer Center Laboratory for Cell Analysis Shared Resource Facility through a grant from the NIH (P30CA082103). We obtained KrasG12D mice from David Tuveson and Tyler Jacks, and Linda Wolff provided the MOL4070LTR retrovirus. This work was supported in part by National Institutes of Health awards R37 CA72614 (KS), R01 CA180037 (KS); P30 CA008748, U54 OD020355 (BST), R01 CA204749 (BST), and awards from the St. Baldrick’s Foundation (KS), the American Cancer Society (RSG-15-067-01-TBG) (KS), the Anna Fuller Fund, and the Robertson Foundation (BST). AMW was supported by a Postdoctoral Fellowship (PF-14-070-01-TBG) from the American Cancer Society including a supplement from the Hillcrest Committee, and an Alex’s Lemonade Stand Foundation Young Investigator Grant including support from Northwestern Mutual. KS is an American Cancer Society Research Professor (RP 10-078-01-TBE).
Author information
Authors and Affiliations
Contributions
AMW, BJH, JWC, SCK, JCA, BST and KS designed experiments and analyzed data. AMW, JWC, KH, HY, LKM, AS, GM and JN performed experiments. BJH, PJ and BST performed bioinformatics analysis. MD, QL, JCW, OW, RPH, SCK, DS and JCA provided reagents. KY, JRD, JZ and MD provided conceptual advice. AMW and KS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Deepak Sampath and Monique Dail were employees and shareholders of Genentech, Inc. when this work was performed. The remaining authors of this paper declare no financial interest related to this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wandler, A.M., Huang, B.J., Craig, J.W. et al. Loss of glucocorticoid receptor expression mediates in vivo dexamethasone resistance in T-cell acute lymphoblastic leukemia. Leukemia 34, 2025–2037 (2020). https://doi.org/10.1038/s41375-020-0748-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-020-0748-6
This article is cited by
-
Exploring the nexus of nuclear receptors in hematological malignancies
Cellular and Molecular Life Sciences (2024)
-
Epigenomic profiling of glucocorticoid responses identifies cis-regulatory disruptions impacting steroid resistance in childhood acute lymphoblastic leukemia
Leukemia (2022)
-
Characterization of a novel glucocorticoid-resistant human B-cell acute lymphoblastic leukemia cell line, with AMPK, mTOR and fatty acid synthesis pathway inhibition
Cancer Cell International (2021)
-
The Aurora kinase/β-catenin axis contributes to dexamethasone resistance in leukemia
npj Precision Oncology (2021)
-
SIRT1 single-nucleotide polymorphisms are associated with corticosteroid sensitivity in primary immune thrombocytopenia patients
Annals of Hematology (2021)