Abstract
The aim of this study was to determine the mechanism by which SIRT6 regulates glucolipid metabolism disorders. We detected histological and molecular changes in Sprague-Dawley rats as well as in BRL 3A and INS-1 cell lines subjected to overnutrition and starvation. SIRT6, SREBP1c, and glucolipid metabolism biomarkers were identified by fluorescence co-localization, real-time PCR, and western blotting. Gene silencing studies were performed. Recombinant SIRT6, AMPK agonist (AICAR), mTOR inhibitor (rapamycin), and liver X receptor (LXR) agonist (T0901317) were used to pre-treated in BRL 3A and INS-1 cells. Real-time PCR and western blotting were used to detect related proteins, and cell counting was utilized to detect proliferation. We obtained conflicting results; SIRT6 and SREBP1c appeared in both the liver and pancreas of high-fat and hungry rats. Recombinant SIRT6 alleviated the decrease in AMPKα and increase in mTORC1 (complex of mTOR, Raptor, and Rheb) caused by overnutrition. SIRT6 siRNA reversed the glucolipid metabolic disorders caused by the AMPK agonist and mTOR inhibitor but not by the LXR agonist. Taken together, our results demonstrate that SIRT6 regulates glycolipid metabolism through AMPKα-mTORC1 regulating SREBP1c in the liver and pancreas induced by overnutrition and starvation, independent of LXR.
Similar content being viewed by others
Introduction
Metabolic syndrome (MetS) is a comprehensive cluster of obesity, dyslipidemia, hyperglycemia, and hypertension caused by energy utilization and storage disorders1, with pathological changes in glucolipid metabolism in multiple organs2. Previous studies have shown the role of Sirtuin 6 (SIRT6) in regulating other lipid metabolic tissues, such as the biliary tract and skeletal muscle, via AMPK alone3,4,5. However, the relationship of SIRT6 via the complete AMPK-mTORC1 pathway in the liver and pancreas has yet to be fully elaborated. SIRT6 is a stress response protein deacetylase and anti-ADP ribose transferase and is involved in a variety of molecular pathways related to aging6. AMP-activated protein kinase α (AMPKα) and mammalian target of rapamycin (mTOR) are serine/threonine protein kinases. The mTOR, Raptor, and Rheb are the constituents of the mTOR complex 1 (mTORC1). AMPKα-mTORC1 are key regulators of metabolism3. The purpose of this study was to explore the specific mechanism by which SIRT6 regulates glucolipid metabolism disorders in the liver and pancreas by detecting histological and molecular changes in models of high-fat diets and starvation in vivo and in vitro.
Materials and methods
Reagents, primers, and antibodies
Small interfering RNA (siRNA) and the negative control and for targeting SIRT6 were designed and synthesized by GenePharma. AMPK agonist (AICAR) and mTOR inhibitor (Rapamycin) were purchased from Cell Signaling Technology. The liver X receptor (LXR) agonist (T0901317) and recombinant SIRT6 were obtained from Abcam. Antibodies were bought from Abcam and Cell Signaling Technology, and primer sequences were designed and synthesized by GenePharma. This information is summarized in Table 1. siRNAs were transfected into cells using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions.
Animal models
Sprague-Dawley rats (SD, male, specific pathogen-free [SPF] grade, 7 weeks old, 180–220 g, Beijing Vital River Laboratory Animal Technology Co., Ltd) were fed at a constant temperature (23 ± 2 °C), humidity (50–60%), and 12 h light/dark cycle with free access to water and food. All experiments were conducted daily from 9 to 11 a.m. to avoid circadian rhythm effects and were approved by the Institutional Animal Care and Use Committee (IACUC) of China Medical University (Approval No. 2020118). After a 1-week adaption, rats were randomly assigned to three groups: normal control (NC, n = 8 rats, fed with a control diet, D12450J, 10:20:20 kcal% of fat, carbohydrate, and protein-energy ratio with a total energy of 3.85 kcal/gm, Research Diets, USA), starvation (ST, n = 8 rats, fed with a control diet, D12450J, 10:20:20 kcal% of fat, carbohydrate and protein-energy ratio with a total energy of 3.85 kcal/gm, Research Diets, USA, starved for three days before the experiment endpoint7), and high fat (HF, n = 8 rats, fed with a high-fat diet, D12492, 60: 20: 20 kcal% of fat, carbohydrate and protein-energy ratio with a total energy of 5.24 kcal/gm, Research Diets, USA).
Cell culture
Rat insulinoma cells (INS-1 cell line, CL-0368, Procell Life Science & Technology) were treated with Roswell Park Memorial Institute (RPMI) 1640 culturing medium with 10% fetal bovine serum (FBS, North American blood source, Gibco) and 50 μM β-mercaptoethanol. Buffalo rat liver cells (BRL 3A cell line, ATCC® CRL-1442) were treated with Dulbecco’s Modified Eagle Medium (DMEM) culturing medium with 10% FBS. Cells were cultured at 37 °C, saturated humidity, in a 5% CO2 incubator, and the medium was changed every day and passed on every other day. After being synchronized via starvation in Opti-MEM (Gibco, USA) for 24 h when the cell confluency reached 70–80%, cells were divided into groups: NC (11.1 mM glucose), ST (free glucose and FBS), and high glucose (HG, 33.3 mM glucose)8 or HF (200 μM palmitic acid)9. After being cultured for 24 h, cells were collected and stored at −80 °C.
Gene silencing and drug treatment
Cells were inoculated into six-well culturing plates and then transfected with SIRT6 and normal control siRNA for gene silencing via Lipofectamine 3000 after 24 h synchronization of Opti-MEM starvation. The recombinant SIRT6 protein (1 μM for 24 h in INS-1 and BRL 3A), AMPK agonist (AICAR, 0.5 mM for 8 h in INS-1 and 0.5 mM for 30 min in BRL 3A), mTOR inhibitor (Rapamycin, 10 µM for 48 h in INS-1 and 20 nM for 24 h in BRL 3A), and LXR agonist (T0901317, 10 μM for 36 h in INS-1 and 200 nM 16 h in BRL 3A) were used to pre-treat cells according to previous studies10,11,12,13,14,15. Then, these cells were collected after 24 h different medium culturing and stored in liquid nitrogen for the follow-up experiment.
Biochemical detection
Rat blood samples were collected by standard sampling. Rat serum liver function and blood lipids were analyzed via a chemiluminescent immunoassay using alanine transaminase (ALT, C009-2-1), aspartate aminotransferase (AST, C010-2-1), triglycerides (TG, A110-1-1), total cholesterol (TC, A111-1-1), low-density lipoprotein cholesterol (LDL-C, A113-1-1), and high-density lipoprotein cholesterol (HDL-C, A112-1-1) kits (Nanjing Jiancheng Bioengineering Institute, China).
mRNA real-time PCR assay
Trizol RNA separation reagent was used to extract total RNA from samples. One g of total RNA was reverse-transcribed into cDNA using a reverse-transcription kit (TaKaRa, Japan). Real-time PCR was conducted with a SYBR® Premix ExTaqTM II kit (TaKaRa, Japan). The reaction system was 20 μl containing 2 μl cDNA. The reaction procedure was as follows: initial denaturation at 95 °C for 3 min, followed by denaturation at 95 °C for 12 s 40 cycles, annealing at 62 °C for 30 s, and elongation at 72 °C for 30 s. At the end of the experiment, the CT value was read for dissolution curve analysis using β-actin (Bact, GenePharma) as a standardized internal reference. The 2−ΔΔCT method was adopted to calculate relative expressions16.
Western blotting
Protein samples were extracted using sample lysates containing protease inhibitors, and total protein was analyzed using a Pierce™ BCA Protein Assay Kit (Thermo Scientific™, USA). The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was conducted after protein denaturing, and then bands were transferred to the PVDF membrane at a specific time. The membrane was incubated with 5% bovine serum albumin (BSA, A1933, Sigma-Aldrich, USA) for 2 h, followed by incubation with primary antibodies at the corresponding concentration at 4 °C overnight. After incubation with the secondary antibody at room temperature for 2 h, ECL luminous fluid (Pierce™ ECL Western Blotting Substrate, Thermo Scientific™, USA) and an imaging system (MicroChemi 4.2, Israel) were adopted to detect the bands, and Fiji software (for macOS, version 2.0.0-rc-69/1.52i with Java 1.8.0_172[64-bit]) was used to detect grey values17.
Cell counting kit-8
A Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was used to analyze the cell survival activity in INS-1 and BRL 3A cell lines treated with different reagents. Cell suspensions were prepared after trypsin digestion, and the cell density was adjusted to 5 × 103/100 μl/well in a 96-well plate with saturated humidity at 37 °C in 5% CO2. Then, 10 μl detection solution was added to each well and incubated for 2 h according to the manufacturer’s instructions. The absorbance at 450 nm wavelength was measured by a full-wavelength microarray (BioTek Power Wave XS, USA) with three wells for each sample.
Double-label immunofluorescence
After fixing in cold acetone, tissues were incubated with primary antibodies (SIRT6 and SREBP1c) at 4 °C overnight. The tissues were then washed and incubated with the secondary antibody at room temperature for 1 h. Sections were washed again and then DAPI stained at room temperature for 10 min. Images of the sections were taken under an Olympus fluorescence confocal microscope (FV-1000, Japan). Caseviewer software (2.3 version 64-bit, 3DHISTECH Ltd.) was used for image collection.
Statistical analysis
Experiments were repeated three times under the same conditions. All obtained data were analyzed with SPSS 20.0 software (IBM, USA). After the normal distribution test in each group, normal distribution data were expressed by the mean ± standard error of the mean, whereas non-normal distribution data were expressed by the median (interquartile range). A Student’s t test for two groups and one-way ANOVA for three groups or more were used to compare the data. The least-squares method t-test was used for comparison between groups with homogeneous variances and Tamhane’s T2 test for different variances. P < 0.05 was considered statistically significant.
Result
Physiological indices of starvation and obesity
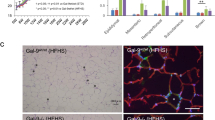
To investigate the role of SIRT6 in energy metabolism, we constructed both energy surplus and deficiency models induced by either a high-fat diet or starvation. Energy-related physiological indicators showed that compared with the normal control group, serum TC, LDL-C, ALT, AST, body weight, kidney weight ratio, body fat ratio (including the omental, perirenal, epididymal, and interscapular fat), liver weight ratio, and liver TG raised in the high-fat group, whereas serum HDL-C decreased (P < 0.05), but there was little change in subcutaneous fat, liver, and pancreas weight ratios, as well as serum TG (P > 0.05). The body weight and body fat ratio were significantly reduced (P < 0.05), but the proportion of each fat component was relatively average, whereas the other indicators showed little change in the starvation group (P > 0.05, Fig. 1a–s).
a fat percentage; b fat ingredients; c body fat ratio; d body weight; e–i perirenal, epididymis, omentum, subcutaneous, and scapular fat ratios; j–n: serum TC, serum TG, liver TG, serum HDL-C, and serum LDL-C levels; o–q pancreas, kidney, and liver ratios; r and s serum ALT and AST levels in three groups; t IPGTT test and area under curve (AUC); u IPITT test and AUC; v HOMA-IR; w ISI; x fast insulin; y and z Acc, Fasn, Ldlr, Pck, and Gck1 levels. *Compared with the NC group, P < 0.05; #Compared with the ST group, P < 0.05. n = 8 per group.
To study changes in glucose metabolism, the results of IPGTT showed that the peak of glucose after glucose injection in both ST and HF groups was delayed to different degrees (the peak of glucose: 5 min in the NC group, 15 min in the ST group, and 30 min in the HF group), and the area under curve (AUC) of IPGTT in both ST and HF groups was significantly increased (P < 0.05, Fig. 1t), indicating high glucose sensitivity in both groups. Moreover, the IPITT experiment showed that glucose levels decreased in all 3 groups after insulin injection, but the decrease was not obvious in the ST group and HF groups compared with the NC group. The AUC in the ST group was significantly lower, and the AUC in the HF group was significantly higher than that of the NC group (P < 0.05, Fig. 1u), indicating high insulin resistance in both ST and HF groups with low and high blood glucose levels. In addition, insulin and HOMA-IR levels were significantly increased, and ISI levels were decreased in the HF group, whereas these levels in the ST group showed opposite results (P < 0.05, Fig. 1v–x), indicating high insulin sensitivity in the ST group and high insulin resistance in the HF group.
Acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN), and low-density lipoprotein receptor (LDLR) are involved in liver lipid metabolism, whereas glucokinase (GCK) and phosphoenolpyruvate carboxykinase 1 (PCK1) are involved in pancreas glucose metabolism. Compared with the NC group, the mRNA levels of liver Acc, Fasn, and Ldlr and pancreas Gck and Pck1 mRNA in the HF group were significantly increased, and these indicators were significantly decreased in the ST group (P < 0.05, Fig. 1y, z).
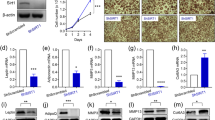
SIRT6 and pathway levels in the status of starvation and obesity
To study the expression of SIRT6 in different energy models, SIRT6 expression in tissues was determined by fluorescence co-localization. SIRT6 in the liver and pancreas of the HF group showed lower expression, but these levels were high in the ST group. Moreover, SIRT6 and SREBP1c have co-location in the pancreas and liver tissues but expressed the opposite (Figs. 2a and 3a). Then, PCR quantitation was used to detect the mRNA expression of Sirt6 in liver and pancreas tissues. The mRNA and protein expression levels of Sirt6 in the HF group were significantly decreased, whereas that of the ST group was significantly increased. The expressions of Ampk and mtor mRNA and related proteins p-AMPKα/AMPKα and mTORC1 complex related factors (p-mTOR/mTOR, p-Raptor/Raptor, and Rheb) were significantly increased in the HF group, but the changes in the ST group showed the opposite (P < 0.05, Figs. 2b–g and 3b–g).
a co-localization of SIRT6 and SREBP1c via fluorescent double staining in liver; b–g levels of SIRT6, p-AMPKα/AMPKα, p-mTOR/mTOR, p-Raptor/Raptor, and Rheb in liver; h–j the abundance of Sirt6, Ampk, and mtor mRNA in liver. *Compared with the NC group, P < 0.05; #Compared with the ST group, P < 0.05. n = 8 per group.
a co-localization of SIRT6 and SREBP1c via fluorescent double staining in pancreas; b–g levels of SIRT6, p-AMPKα/AMPKα, p-mTOR/mTOR, p-Raptor/Raptor, and Rheb in pancreas; h–j the abundance of Sirt6, Ampk, and mtor mRNA in pancreas. *Compared with the NC group, P < 0.05; #Compared with the ST group, P < 0.05. n = 8 per group.
Changes in free and high-fat BRL 3A and INS-1 cells
To investigate the regulation mechanism of SIRT6 in glucolipid metabolism of the liver and pancreas, we constructed liver BRL 3A cell lines cultured with high fat and starvation as well as pancreatic INS-1 cell lines induced by high glucose and starvation, respectively. It was found that in BRL 3A cells, the mRNA expressions of Sirt6, Ampk, and mtor, the related proteins SIRT6, p-AMPKα/AMPKα, and the mTORC1 complex-related factors (p-mTOR/mTOR, p-Raptor/Raptor, and Rheb) were significantly increased, the mRNA expressions of liver metabolic enzymes (Acc, Fasn, and Ldlr) were significantly increased, and the activity of CCK-8 was significantly decreased in the HF group. The changes of the ST group provided the opposite results (P < 0.05, Fig. 4). In the INS-1 cells, the mRNA expressions of Sirt6, Ampk, and mtor, the related proteins SIRT6, p-AMPKα/AMPKα, and the mTORC1 complex-related factors (p-mTOR/mTOR, p-Raptor/Raptor, and Rheb) were significantly increased, the mRNA expressions of liver metabolic enzymes (Gck and Pck1) were significantly increased, and the activity of CCK-8 was significantly decreased in the HG group. The changes of ST and HG groups were found to be the opposite (P < 0.05, Fig. 5).
a cell viability tested by CCK-8; b–d metabolic enzyme abundance of Acc, Fasn, and Ldlr; e–g the abundance of Sirt6, Ampk, and mtor mRNA; h–m: levels of SIRT6, p-AMPKα/AMPKα, p-mTOR/mTOR, p-Raptor/Raptor, and Rheb in BRL 3A cells. *Compared with the NC group, P < 0.05; #Compared with the ST group, P < 0.05. n = 6 per group.
a cell viability tested by CCK-8; b and c metabolic enzyme abundance of Gck and Pck1; d–f the abundance of Sirt6, Ampk, and mtor mRNA; g–l levels of SIRT6, p-AMPKα/AMPKα, p-mTOR/mTOR, p-Raptor/Raptor, and Rheb in INS-1 cells. *Compared with the NC group, P < 0.05; #Compared with the ST group, P < 0.05. n = 6 per group.
SIRT6 regulating AMPKα-mTORC1 pathway in BRL 3A and INS-1 cells
To explore the role of SIRT6 regulating AMPKα-mTORC1, we pre-treated cells with recombinant human SIRT6 protein, SIRT6 siRNA, AMPK agonist (AICAR), and mTORC1 inhibitor (rapamycin), and cultured cells with normal and high glucose or fat media, and pathway-related mRNA and protein were detected. The results showed that recombinant human SIRT6 protein could reverse glucolipid metabolic changes in BRL 3A and INS-1 cells pre-treated with the high glucose and fat media via the AMPKα-mTORC1 pathway (Supplementary Fig. 1), and SIRT6 siRNA was able to reverse the changes of AMPK agonist and mTOR inhibitors, respectively (Supplementary Figs. 2 and 3). Various metabolic enzymes (Acc, Fasn, and Ldlr mRNA in BRL 3A cells and Gck and Pck1 mRNA in INS-1 cells) and CCK-8 cell vitality all changed with these pre-treatments. Srebp1c mRNA plays a role in glucolipid metabolism via transcription, processing, and nSREBP1c nuclear import by the regulation of SREBP cleavage-activating protein (SCAP), Insig 1, Insig 2, and Lipin-1. Here, we found that the AMPK agonist (AICAR) could regulate Lipin-1, mTOR inhibitor (rapamycin) could regulate the entire pathway, and SIRT6 siRNA was able to reverse the corresponding effect of SREBP1c produced in both pre-treated BRL 3A and INS-1 cells (Supplementary Figs. 2 and 3).
Regulating ability of SIRT6 is independent of LXR
Previous studies have shown that changes in glucolipid metabolism mediated by SREBP1c may be regulated by LXR. To explore the relationship between SIRT6 and LXR, SIRT6 siRNA, and LXR agonist (T0901317) were used to pre-treat BRL 3A and INS-1 cells, and the levels of pathway-related factors were detected. The results showed that both SIRT6 siRNA and LXR agonist (T0901317) could lead to changes of various metabolic enzymes and cell phenotypes in BRL 3A and INS-1 cells; however, they had no regulatory effect on each other (Supplementary Fig. 4), suggesting that the role of SIRT6 in regulating SREBP1c-mediated glucolipid metabolism may be independent of LXR.
Discussion
MetS is a clustering of medical conditions comprising obesity, dyslipidemia, hyperglycemia, hypertension, and other additional components and is caused by an underlying disorder of energy utilization and storage1. Its high prevalence and large number of complications have drawn people’s attention in recent years. The most basic pathological change of MetS is glucolipid metabolism in multiple organs2. A successful model of diabetes and obesity established rat high-glycemic islet cells in our previous research showed the relationship between MetS and diabetes by regulating glucose metabolism and insulin resistance18, 19. SIRT6 is a stress-responsive protein deacetylase and mono-ADP ribosyl transferase enzyme, the functions of which are associated with multiple molecular pathways related to aging, including DNA repair, telomere maintenance, glycolysis, and inflammation3. On the basis of previous research, this study established rat models of obesity and starvation, and cultured islet and liver cell lines successfully. It was found that the expression of SIRT6 was significantly changed in vivo and in vitro in the state of obesity and starvation, suggesting that SIRT6 may be related to the energy balance of obesity and starvation.
Energy balance is regulated by AMPK, a silk/threonine protein kinase, at the overall level. The α subunit of AMPK contains kinase activity, and phosphorylation of the 172nd threonine (T172) is the key to activation20. Glucose starvation is directly sensed and causes AMPK activation21, 22. The mTOR is also a silk/threonine protein kinase related to the signaling pathways of growth, nutrition, and energy supply. The complex mTORC1, composed of mTOR, Raptor, and Rheb, is a rapamycin-sensitive protein that regulates cell growth through protein translation and is associated with a variety of metabolic diseases, such as diabetes20. mTORC1 is activated in the case of abundant nutrients, which accelerates cell growth by promoting anabolism23, whereas AMPK can directly inhibit the activity of Rheb and mTORC1 by phosphorylating TSC2 or directly phosphorylate Raptor in the mTORC1 complex to cause changes in the structure of the mTORC1 complex20, 24. Besides, AMPK can further phosphorylate leucoamanyl-tRNA synthase, LARS1, by activating downstream ULK1 to regulate mTORC125. At the same time, the active mTORC1 can further inhibit AMPK with low vitality under sufficient nutrition conditions26. This evolutionary “double insurance” ensures that AMPKα-mTORC1 can maintain the balance with the change of nutrients or energy, indicating that AMPKα-mTORC1, as the “Yin and Yang” sides of metabolic regulation, also has a mutual regulatory effect, encouraging us to update the current SIRT6 research.
Previous studies have shown that SIRT6 inhibits glycochenodeoxycholate-induced biliary epithelial cell apoptosis by the AMPK/peroxlsome proliferator-activated receptor-γ coactlvator-1α pathway4 and regulates skeletal muscle metabolic homeostasis by activating AMPK6, suggesting that SIRT6 regulates other lipid metabolic tissues via AMPK alone. To investigate the specific regulation of SIRT6 on the AMPKα-mTORC1 pathway in glucolipid metabolic tissues, particularly for the detection of the mTORC1 complex comprising mTOR, Raptor, and Rheb, we used recombinant human SIRT6 protein, SIRT6 siRNA, AMPKα agonist, and mTORC1 inhibitor for cell pre-treatment. The results showed that SIRT6 siRNA and recombinant protein could change the expression levels of phosphorylated AMPKα and mTORC1 (Rheb, phosphorylated Raptor, and mTOR in the complex), whereas the AMPKα agonist and mTOR inhibitor only interacted with each other but could not change SIRT6 levels, suggesting that SIRT6 has a regulatory effect on the complete AMPKα-mTORC1 pathway in the pancreas and liver.
SREBP1 is an important nuclear transcription factor of lipid metabolism, mainly regulating the synthesis of fatty acids, triglycerides, and cholesterol, and is closely associated with MetS-related diseases such as insulin resistance, diabetes, and fatty liver. Its isoform SREBP1c, specifically expressed in the liver, plays a key role in the formation of triglycerides and phospholipids27,28,29, the related glycolipid metabolic enzymes of which include ACC, FASN, and LDLR in lipid metabolism and GCK and PCK1 in glucose metabolism30, 31. Studies have shown that SIRT6 plays a role in cholesterol regulation through the lipid-generating transcription factors SREBP1 and 2 introns by AMPK5. Moreover, SIRT6 activates the PPARα and its dependent pathway by NCOA2 deacetylation and regulates PPARα and SREBP1c-dependent cholesterol synthesis and fatty acid oxidation in a variety of multiple ways, including mitochondrial oxidation, peroxidosomal oxidation, acyl coenzyme A binding or hydrolysis, lipid storage or transport, and ketone production32, indicating that SIRT6 regulates lipid metabolic tissues by SREBP1c. To further study the specific mechanism of SIRT6 regulating SREBP1c throughout the entirety of glucolipid metabolic tissues, we detected the genes regulated by SREBP1c and found that the mRNA levels of Acc, Fasn, and Ldlr, in liver tissues and Gck and Pck1 in pancreas tissues were significantly changed by SIRT6, suggesting that SIRT6 was significantly related to glucolipid metabolism mediated by SREBP1c.
Sterol regulatory element-binding protein 1 (SREBP1), specifically expressed in the liver as the SREBP1c isoform, is an important nuclear transcription factor in glucolipid metabolism, is closely associated with a variety of MetS-related diseases33, and has been found to be regulated by LXR in previous studies34,35,36. SREBP1c exists in the endoplasmic reticulum of cells, and only after proteolytic nucleation can it become a mature karyotype (nSREBP1c) and play a role in transcriptional regulation downstream30. Insig1 and Insig2 are endoplasmic reticulum proteins that block the processing of SREBP1c by binding to SCAP, thus preventing SCAP from escorting SREBPs to the Golgi37. AMPK can prevent nSREBP1c from entering the nucleus to affect lipid metabolism, whereas insulin can regulate SREBP1 transcription through the PI3K/AKT/mTORC1 pathway, and mTORC1 can affect the processing of SREBP1c through S6K and the formation of nSREBP1c through Lipin-1, thus promoting the development of NAFLD33, 38, 39.
To explore the specific regulatory mechanism of SIRT6 regulating SREBP1c, the SIRT6 siRNA, AMPK agonist, and mTOR inhibitor were adopted, and the changes of downstream effector molecules (AMPKα-mTORC1 and glycolipid metabolic enzymes) were detected in each stage of the pathway. The results showed that SIRT6 could reverse the effects of AMPK and mTOR inhibitors regulating the transcription, processing, and nucleation of SREBP1c, but usage of the LXR agonist T0901317 did not reverse these changes, suggesting that SIRT6 regulates the metabolism of SREBP1c through the AMPKα-mTORC1 pathway and then affects glucolipid metabolic enzymes in an LXR-independent pathway.
In conclusion, SIRT6 regulates glycolipid metabolism through AMPKα-mTORC1 regulating SREBP1c in the liver and pancreas induced by overnutrition and starvation, independently of LXR. The specific molecular mechanism is shown in Fig. 6.
Only after the proteolytic nucleation processing in endoplasmic reticulum can SREBP1 (specifically expressed SREBP1c in liver) become a mature karyotype nSREBP1c participating in downstream transcription. Insig1 and Insig2 block SREBP1c processing by binding to SCAP, thus preventing SCAP from escorting SREBPs to the Golgi. AMPK can prevent nSREBP1c from entering the nucleus, whereas mTORC1 can affect SREBP1c processing and nSREBP1c formation through Lipin-1. Previously, SREBP1c is regulated by LXR in the classical pathway. Here, we found that SIRT6 regulates glycolipid metabolism (ACC, FASN, and LDLR for lipid metabolism in liver; GCK and PCK1 in glucose metabolism in pancreas) through the AMPKα-mTORC1 complex (T172 p-AMPKα, mTOR, Raptor, and Rheb) regulating SREBP1c in the liver and pancreas induced by overnutrition and starvation, independently of LXR.
Ethics approval
All animals were kept in a pathogen-free environment and fed ad libitum. The procedures for care and use of animals were approved by the IACUC of the China Medical University (Approval No. 2020118), and all experimental operations complied with the Guide for Laboratory Animal Care and Use and Animal Welfare Act. All applicable institutional and governmental regulations concerning the ethical use of animals were followed.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Beltran-Sanchez, H., Harhay, M. O., Harhay, M. M. & McElligott, S. Prevalence and trends of metabolic syndrome in the adult U.S. population, 1999-2010. J. Am. Coll. Cardiol. 62, 697–703 (2013).
Priest, C. & Tontonoz, P. Inter-organ cross-talk in metabolic syndrome. Nat. Metab. 1, 1177–1188 (2019).
Li, J. et al. Sirt6 opposes glycochenodeoxycholate-induced apoptosis of biliary epithelial cells through the AMPK/PGC-1alpha pathway. Cell Biosci. 10, 43 (2020).
Elhanati, S. et al. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 4, 905–912 (2013).
Cui, X. et al. SIRT6 regulates metabolic homeostasis in skeletal muscle through activation of AMPK. Am. J. Physiol. Endocrinol. Metab. 313, E493–E505 (2017).
Frye, R. A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 (2000).
Boland, B. B. et al. beta-Cell control of insulin production during starvation-refeeding in male rats. Endocrinology 159, 895–906 (2018).
Li, Q., Jia, S., Xu, L., Li, B. & Chen, N. Metformin-induced autophagy and irisin improves INS-1 cell function and survival in high-glucose environment via AMPK/SIRT1/PGC-1alpha signal pathway. Food Sci. Nutr. 7, 1695–1703 (2019).
Zhang, Y. et al. Silibinin ameliorates steatosis and insulin resistance during non-alcoholic fatty liver disease development partly through targeting IRS-1/PI3K/Akt pathway. Int. Immunopharmacol. 17, 714–720 (2013).
Klein, M. A. et al. Mechanism of activation for the sirtuin 6 protein deacylase. J. Biol. Chem. 295, 1385–1399 (2020).
Guo, H. et al. AMPK enhances the expression of pancreatic duodenal homeobox-1 via PPARalpha, but not PPARgamma, in rat insulinoma cell line INS-1. Acta Pharm. Sin. 31, 963–969 (2010).
Ren, G., Rimando, A. M. & Mathews, S. T. AMPK activation by pterostilbene contributes to suppression of hepatic gluconeogenic gene expression and glucose production in H4IIE cells. Biochem. Biophys. Res. Commun. 498, 640–645 (2018).
Mwangi, S. M. et al. Glial cell line-derived neurotrophic factor enhances autophagic flux in mouse and rat hepatocytes and protects against palmitate lipotoxicity. Hepatology 69, 2455–2470 (2019).
Choe, S. S. et al. Chronic activation of liver X receptor induces beta-cell apoptosis through hyperactivation of lipogenesis: liver X receptor-mediated lipotoxicity in pancreatic beta-cells. Diabetes 56, 1534–1543 (2007).
Comas, J. & Vives-Rego, J. Assessment of the effects of gramicidin, formaldehyde, and surfactants on Escherichia coli by flow cytometry using nucleic acid and membrane potential dyes. Cytometry 29, 58–64 (1997).
Karatayli, E., Hall, R. A., Weber, S. N., Dooley, S. & Lammert, F. Effect of alcohol on the interleukin 6-mediated inflammatory response in a new mouse model of acute-on-chronic liver injury. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 298–307 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Bian, C., Bai, B., Gao, Q., Li, S. & Zhao, Y. 17beta-Estradiol regulates glucose metabolism and insulin secretion in rat Islet beta cells through GPER and Akt/mTOR/GLUT2 pathway. Front. Endocrinol. 10, 531 (2019).
Ren, H. et al. Metformin alleviates oxidative stress and enhances autophagy in diabetic kidney disease via AMPK/SIRT1-FoxO1 pathway. Mol. Cell Endocrinol. 500, 110628 (2020).
Gonzalez, A., Hall, M. N., Lin, S. C. & Hardie, D. G. AMPK and TOR: the Yin and Yang of cellular nutrient sensing and growth control. Cell Metab. 31, 472–492 (2020).
Zhang, C. S. et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 (2017).
Li, M. et al. Transient receptor potential V channels are essential for glucose sensing by aldolase and AMPK. Cell Metab. 30, 508–524 (2019). e512.
Gonzalez, A. & Hall, M. N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36, 397–408 (2017).
Gwinn, D. M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008).
Yoon, I. et al. Glucose-dependent control of leucine metabolism by leucyl-tRNA synthetase 1. Science 367, 205–210 (2020).
Ling, N. et al. mTORC1 directly inhibits AMPK to promote cell proliferation under nutrient stress. Nat. Metab. 2, 41–49 (2020).
Alves-Bezerra, M. & Cohen, D. E. Triglyceride metabolism in the liver. Compr. Physiol. 8, 1–8 (2017).
Santos-López, J. A., et al. Lipoprotein profile in aged rats fed Chia oil- or hydroxytyrosol-enriched pork in high cholesterol/high saturated fat diets. Nutrients 10, 1830 (2018).
Rohrbach, T. D. et al. FTY720/fingolimod decreases hepatic steatosis and expression of fatty acid synthase in diet-induced nonalcoholic fatty liver disease in mice. J Lipid Res. 60, 1311–1322 (2019).
Bai, Y., et al. Chitosan oligosaccharides improve glucolipid metabolism disorder in liver by suppression of obesity-related inflammation and restoration of peroxisome proliferator-activated receptor gamma (PPARgamma). Mar. Drugs 16, 455 (2018).
Nie, H. et al. O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat. Commun. 11, 36 (2020).
Naiman, S. et al. SIRT6 promotes hepatic beta-oxidation via activation of PPARalpha. Cell Rep. 29, 4127–4143 (2019). e4128.
Georgila, K., Gounis, M., Havaki, S., Gorgoulis, V. G. & Eliopoulos, A. G. mTORC1-dependent protein synthesis and autophagy uncouple in the regulation of Apolipoprotein A-I expression. Metabolism 105, 154186 (2020).
Su, W. et al. Liver X receptor alpha induces 17beta-hydroxysteroid dehydrogenase-13 expression through SREBP-1c. Am. J. Physiol. Endocrinol. Metab. 312, E357–E367 (2017).
Lin, Y. N. et al. Oleanolic acid inhibits liver X receptor alpha and pregnane X receptor to attenuate ligand-induced lipogenesis. J. Agric. Food Chem. 66, 10964–10976 (2018).
Lin, Y. N. et al. Ursolic acid, a novel liver X Receptor alpha (LXRalpha) antagonist inhibiting ligand-induced nonalcoholic fatty liver and drug-induced lipogenesis. J. Agric. Food Chem. 66, 11647–11662 (2018).
Xu, D. et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 580, 530–535 (2020).
Ni, W. et al. USP7 mediates pathological hepatic de novo lipogenesis through promoting stabilization and transcription of ZNF638. Cell Death Dis. 11, 843 (2020).
Hao, E. Y. et al. The relationship between the mTOR signaling pathway and ovarian aging in peak-phase and late-phase laying hens. Poult. Sci. 100, 334–347 (2021).
Funding
This study was supported by the Doctoral Research Initiation Fund Project of Liaoning Province (Grant No. 2021-BS-206) and Shenyang Young and Middle-aged Innovation Support Program (RC210460).
Author information
Authors and Affiliations
Contributions
Che Bian: conceptualization, methodology, formal analysis, and original draft writing. Haibo Zhang and Jing Gao: methodology, formal analysis, and revision. Yuxia Wang, Jia Li, Dan Guo, Wei Wang, Yuling Song, and Yang Weng: methodology and formal analysis. Huiwen Ren: supervision, conceptualization, methodology, software, revision, and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bian, C., Zhang, H., Gao, J. et al. SIRT6 regulates SREBP1c-induced glucolipid metabolism in liver and pancreas via the AMPKα-mTORC1 pathway. Lab Invest 102, 474–484 (2022). https://doi.org/10.1038/s41374-021-00715-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00715-1