Abstract
Determination of 1p/19q co-deletion status is important for the classification, prognostication, and personalized therapy in diffuse lower-grade gliomas (LGG). We developed and validated a deep learning imaging signature (DLIS) from preoperative magnetic resonance imaging (MRI) for predicting the 1p/19q status in patients with LGG. The DLIS was constructed on a training dataset (n = 330) and validated on both an internal validation dataset (n = 123) and a public TCIA dataset (n = 102). The receiver operating characteristic (ROC) analysis and precision recall curves (PRC) were used to measure the classification performance. The area under ROC curves (AUC) of the DLIS was 0.999 for training dataset, 0.986 for validation dataset, and 0.983 for testing dataset. The F1-score of the prediction model was 0.992 for training dataset, 0.940 for validation dataset, and 0.925 for testing dataset. Our data suggests that DLIS could be used to predict the 1p/19q status from preoperative imaging in patients with LGG. The imaging-based deep learning has the potential to be a noninvasive tool predictive of molecular markers in adult diffuse gliomas.
Similar content being viewed by others
Introduction
Diffuse lower-grade gliomas (LGG) are infiltrative neoplasms comprising of world health organization (WHO) Grade II and III gliomas1. The 2016 WHO classification of central nervous system tumors recognized three subtypes of LGG based on histological and molecular characteristics: (1) isocitrate dehydrogenase (IDH) mutant, 1p/19q co-deleted oligodendroglioma, (2) IDH mutant, 1p/19q intact diffuse astrocytoma, (3) IDH wild-type diffuse astrocytoma2. Furthermore, the latest 2021 WHO criteria advances the role of molecular diagnostics in central nervous system tumor classification3. According to this criteria, 1p/19q status is vital to the determination of oligodendroglioma. Moreover, 1p/19q co-deletion has been demonstrated as a favorable prognostic biomarker in LGG due to its predictive value for better response to adjuvant chemotherapy4. Hence, determination of 1p/19q status is important for the classification, prognostication, and personalized therapy in LGG.
Currently, detecting 1p/19q status requires glioma tissues obtained via surgical resection or biopsy followed by fluorescence in-situ hybridization (FISH) to identify chromosomal deletion5. On the other hand, noninvasive detection of 1p/19q status in LGG is challenging. In recent years, advances in artificial intelligence have led to considerable interest in image-based molecular profiling of gliomas6. A few studies utilized handcrafted radiomic features extracted from preoperative magnetic resonance image (MRI) to predict 1p/19q status in LGG via machine learning7,8,9,10. However, handcrafted features rely on current understanding of imaging, therefore its prediction performance may be limited. Unlike handcrafted radiomics, deep learning can automatically learn predictive feature from images by its own11. Recent studies have shown that deep learning approaches have achieved better performance in tumor classification tasks, compared with radiomics and even human experts12,13,14. However, few studies have investigated the feasibility of deep learning in predicting glioma 1p/19q status from imaging, except one most recent study reporting a MRI-based deep learning model built using 368 patients with WHO II-IV gliomas from public the cancer imaging archive (TCIA) database15. However, this study only used T2-weighted MRI in model building, and the obtained model was not tested independently, so its reproducibility was unknown.
In this study including in total of 555 patients, we developed and validated, both internally and externally, a deep convolutional neural network (CNN) built from preoperative multiparametric MRI to predict the 1p/19q status in patients with LGG.
Materials and methods
Patients and datasets
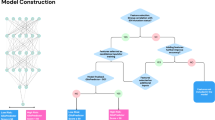
This study was a part of the registered clinical trial “MR Based Prediction of Molecular Pathology in Glioma Using Artificial Intelligence” (ClinicalTrials.gov ID: NCT04217018). This study was approved by the Human Scientific Ethics Committee of the First Affiliated Hospital of Zhengzhou University (No. 2019-KY-176). Informed consents were waived by the Committee due to the retrospective and anonymous nature. There were two datasets included in this study: Dataset 1 contained 453 patients from the first affiliated hospital of zhengzhou university (FAHZZU), Dataset 2 contained 102 patients from TCIA (https://www.cancerimagingarchive.net/). Dataset 1 was divided into two sets: a (1) training set (n = 330) used to develop the deep learning imaging signature (DLIS), and an (2) internal validation set (n = 123) used to optimize the deep CNN. Dataset 2 was used for external validation for the DLIS, as illustrated in Fig. 1. Note that the training and internal validation sets were selected from the FAHZZU cohort using stratified random sampling, where the clinical parameters among these cohorts were balanced. The inclusion criteria are as follows: (1) adult patients (>18 years) surgically treated and pathologically diagnosed as primary WHO II or III diffuse gliomas, (2) availability of clinical data and 1p/19q co-deletion status, (3) availability of preoperative MRI including four sequences: T1-weighted, T2-weighted, T2-weighted fluid-attenuated inversion recovery (FLAIR), and T1-weighted gadolinium contrast-enhanced (T1w, T2w, FLAIR, and T1c), (4) availability of sufficient image quality without significant artifacts, determined by neuroradiologists and neurosurgeons. The selection pipeline for the two datasets is described in Fig. 2. Clinical parameters, including gender, age, and preoperative Karnofsky performance status (KPS) scale were collected from the medical record system of FAHZZU and the cancer genome atlas (TCGA, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) database.
The finally included datasets were the Dataset 1 (FAHZZU) of 453 patients used for model training and internal validation, and the Dataset 2 (TCIA) of 102 patients used for external validation. LGG is short for lower-grade gliomas, FAHZZU is short for the first affiliated hospital of zhengzhou university, TCGA is short for the cancer genome atlas, TCIA is short for the cancer imaging archive, and MRI is short for magnetic resonance imaging.
Detection of chromosome 1p/19q co-deletion status by FISH
Formalin-fixed, paraffin embedded tissues of the FAHZZU dataset were used for detection of chromosome 1p/19q status by FISH. The detailed protocol is described in Supplementary A116.
MRI acquisition
Patients in Dataset 1 (FAHZZU) and Dataset 2 (TCIA) were all performed on either 1.5T or 3.0T clinical MR scanners with different manufacturers and imaging parameters. The protocol in the two datasets all consisted of the following sequences: (a) axial T1w before and after intravenous administration of the gadolinium-based contrast agent (T1c); (b) axial T2w; (c) axial FLAIR. Detailed information about the MR machines and imaging parameters of the two datasets are available in Supplementary A2.
Image preprocessing
A MRI preprocessing pipeline was performed to normalize the image variations across patients. First, N4ITK was applied for bias field distortion correction. After skull stripping, all voxels were isotropically resampled into 1 × 1 × 1 mm3 with linear interpolation for consistent feature extraction. Rigid registration was performed to align the four sequences using T1c as a template with the mutual information similarity metric, generating four registered images rT1w, rT2w, rFLAIR, and rT1c. Tumor contours were manually traced slice-by-slice by a board-certified neuroradiologist (J.Y. with 11 years’ experience) blinded to clinical data in the axial section using the ITK-SNAP software. The whole tumor region, including the contrast-enhancing, nonenhancing, necrosis and edema, if any, was delineated as the signal abnormal regions in the white matter on rFLAIR images. rT2w and rT1c images were used to cross-check the extension of the whole tumor areas. The delineated contours were confirmed by another neurosurgeon (Z.Y.Z. with 11 years’ experience), where uncertainty or disagreement was settled via discussion. Finally, consensus delineation was achieved for each patient.
Deep CNN development
Based on the tumor contours, a 3D bounding box containing the entire tumor was calculated for each patient. To represent the entire tumor, 5 equally-spaced axial slices within the tumor volume were selected from each of the 4 registered MRIs. Then, 20 slices per patient were obtained and used as a single sample of a 3D tumor. Using the 3D bounding box, the selected slices from each MR sequence were cropped into small ones. All slices were resized to 256 × 256. The 20 resized slices were fed into the network as a single input and generated one single output. The deep CNN used for 1p/19q status prediction employed the ResNet-34 architecture as the backbone. The network was trained from scratch on the training set (6600 images, 330 patients) and tuned on the internal validation set (2460 images, 123 patients). During training, an Adam optimizer was used with a learning rate of 0.001 and a batch size of 32. Random rotation, shear and zoom approaches were used for data augmentation. The network output was the DLIS used for predicting the 1p/19q status. Detailed information of the network is described in Supplementary A3.
Statistical analysis
Statistical analysis was performed using R version 3.6.1. The patient and tumor characteristics between training, validation, and testing cohorts were assessed by Wilcoxon test or Chi-square test, where P value <0.05 was considered significant. The performance of the presented deep CNN was independently assessed on the external testing dataset. The receiver operating characteristic (ROC) analysis was used to measure the classification performance in terms of area under the ROC curve (AUC), accuracy, sensitivity, and specificity. The optimal cutoff was chosen by using the X-tile tool. The precision recall curves (PRC) and F1-score were also calculated to evaluate the prediction performance. All indices were calculated for training, internal validation, and testing datasets. For comparison, another two parameters, T2-FLAIR mismatch (yes or not) and T1-contrast enhancement (no, blurry, nodular, and ring-lik enhancement), were calculated from MRI for each patient. First, Wilcoxon test was performed on the entire dataset to evaluate the univariable association of each parameter with 1p/19q co-deletion status, where P < 0.05 indicated the statistical significance. Then, a multivariable model was built by combing the CNN-generated risk score, the T2-FLAIR mismatch, and the T1-contrast enhancement using logistic regression. The performance of the multivariable model was assessed by using ROC analysis.
Results
Patient characteristics
In total 555 patients from FAHZZU and the public database TCIA were enrolled in this study (mean age, 44.6 years ± 12.0 [standard deviation]; 306 male), as summarized in Table 1. The study cohort comprised a training dataset (n = 330, from FAHZZU, mean age, 44.7 years ± 11.6; 193 male), an internal validation dataset (n = 123, from FAHZZU, mean age, 43.1 years ± 11.4; 67 male), and an external testing dataset (n = 102, from TCIA, mean age, 46.3 years ± 13.9; 46 male). As shown in Supplementary Fig. S1, 1p/19q co-deletion was significantly correlated with the overall survival in the training and validation datasets, respectively (P < 0.05). And the 5-year survival rate of patients with 1p/19q co-deletion was significantly higher (P < 0.05).
Classification performance
The ROC curves of the deep learning model for training, internal validation, and testing datasets are shown in Fig. 3A–C. The AUC of the deep learning model was 99.99% for training dataset, 98.62% for validation dataset, and 98.30% for testing dataset. The optimal cutoff determined by using the X-tile tool was 0.65. At this cutoff, the accuracy was 99.30% for training dataset, 94.51% for validation dataset, and 93.33% for testing dataset. The PRC curves of the deep learning model for training, validation, and testing datasets are shown in Fig. 3D–F. The F1-score of the deep learning model was 99.20% for training dataset, 93.99% for validation dataset, and 92.54% for testing dataset. The classification results of the deep learning model are summarized in Table 2. The predictive power of T2-FLAIR mismatch and T1-contrast enhancement was also assessed. The univariable Wilcoxon P value was 0.0003 for T2-FLAIR mismatch and 0.0088 for T1-contrast enhancement, respectively. The multivariable model combining the deep learning risk score, the T2-FLAIR mismatch and the T1-contrast enhancement were built. The multivariable linear model generated by the logistic regression is shown in Supplementary A4, where the weight of the deep learning risk score was larger than that of the other two parameters. The AUC of the multivariable model was 99.0% for training dataset, 99.9% for validation dataset, and 99.9% for testing dataset. DeLong analysis reveals that there was no significant difference between the AUCs of the deep learning model and the multivariable model on each dataset. The ROC curves and the classification performance of the multivariable model are shown in Supplementary Fig. S2 and Supplementary Table S1.
Interpretation of the deep CNN prediction
To illustrate where the deep learning model focused on for 1p/19q status prediction, the class activation maps (CAMs) of the deep CNN with corresponding MRI of four representative patients classified into 1p/19q co-deleted and 1p/19q intact groups are shown in Fig. 4. These maps show that deep CNN could highlight regions relevant to prediction while suppress other less relevant regions. This heatmap-like display allows assessing the tumoral regions with potential predictive value. To further illustrate the relevance of the deep CNN features with the 1p/19q status, a heatmap of the 1p/19q status-specific features is presented in Fig. 5. The deep features in this heatmap were extracted from the fully connected layer used for final prediction.
Discussion
In this retrospective multi-center study, we developed and validated, both internally and externally, a deep learning model predictive of 1p/19q co-deletion status from preoperative MRI in patients with LGG. Aside from its diagnostic and prognostic values, 1p/19q status has also been revealed as an informative biomarker to surgical plans in LGG17,18,19,20,21. More specifically, studies have showed postsurgical residual tumor has a more negative impact on the survival of the IDH mutant, 1p/19q intact gliomas than that of the IDH mutant, 1p/19q co-deleted gliomas18,19. Another study suggested 1p/19q intact but not 1p/19q co-deleted LGG would benefit from gross total resection (GTR)20. It was also reported that the survival difference was nonsignificant between GTR and non-GTR in 1p/19q co-deleted WHO III gliomas, but this difference was significant in 1p/19q intact WHO III gliomas21. However, in current clinical scenario, determining 1p/19q status of gliomas necessitates an invasive surgical resection or biopsy. Hence, preoperative noninvasive detection of 1p/19q status would better aid neurosurgeon to make an informed surgical plan, avoiding non-maximal resection in 1p/19q intact LGG or less-justified surgery-related neurological deficits in 1p/19q co-deleted LGG.
Recent imaging-genomic, or radiogenomic studies revealed the potential link between imaging traits and underlying genetic alterations. Previously, a visual-based MRI characteristic, T2-FLAIR mismatch sign was recognized a specific imaging biomarker that may discriminate 1p/19q status in LGG22. Nevertheless, the sensitivity of T2-FLAIR mismatch sign was too low (30–34%)23, which may hamper its utility in clinical application. On the other hand, recent advanced in medical image analysis allows us to extract numerous quantitative features from imaging. These imaging features can be used to predict clinical outcomes or molecular characteristics via machine learning approach. Several studies focused on handcrafted radiomics features extracted from MRI to predict 1p/19q status and reached AUCs no more than 0.9, with only one study including an independent set for model validation7,8,9,10. More recently, Yogananda et al developed a deep learning model using only T2-weighted MR images to predict 1p/19q status in TCIA database15, where deep features from multiparametric MRI such as T1w, FLAIR, and T1c were not investigated.
Our deep learning algorithm was trained on 330 patients (AUC 0.999), validated in 123 patients (AUC 0.986), and tested in an independent TCIA set (AUC 0.983). Compared with previous 1p/19q prediction models based on handcrafted radiomic or deep learning7,8,9,10,11, our model reached the highest accuracy, sensitivity, and specificity. The reasons for this phenomenon may be the large amount of sample size and the inclusion of multiparametric MRI (T1w, T2w, FLAIR, and T1c) for training the deep learning model. In addition, the performance of our model was tested in a multi-institutional TCIA set and reached a considerably high AUC, which demonstrated the generalizability of our model. Furthermore, we demonstrate the localizability of the CAMs of deep features in our approach for 1p/19q status prediction. Figure 4 shows the example of CAMs of the DLIS in two grade II/III 1p/19q co-deleted and two grade II/III 1p/19q intact patients. These heatmaps illustrate that the deep CNN model pays most attention to the contrast-enhancing, nonenhancing, and necrosis areas, while they ignores the edema areas included in the whole tumor regions. Thus, we deduce that the contrast-enhancing, nonenhancing, and necrosis areas in the whole tumor regions have the most potential predictive value.
Despite the encouraging results, several limitations should be noted. First, our study only includes the conventional MR sequences, incorporating advanced MRI sequences such as diffusion and perfusion MRI may provide additional information and boost prediction performance for 1p/19q status24,25. Second, deep learning image features excavated by black-box-like networks are vast, nameless, and obscure, which is a major obstacle lies in the way of translating deep learning prediction model into clinical practice. More co-clinical and pre-clinical experimental work should be warranted to clarify the biological basis of our presented model.
In conclusion, our findings demonstrate that a deep CNN model built from preoperative multiparametric MRI could predict the 1p/19q status in LGG patients with high accuracy, sensitivity, and specificity. The imaging-based deep learning has the potential to be a noninvasive tool predictive of vital molecular markers in adult diffuse gliomas.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Brat, D. J. et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl. J. Med. 372, 2481–2498 (2015).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta. Neuropathol. 131, 803–820 (2016).
Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 23, 1231–1251 (2021).
Kaloshi, G. et al. Temozolomide for low-grade gliomas: predictive impact of 1p/19q loss on response and outcome. Neurology. 68, 1831–1836 (2007).
Scheie, D. et al. Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. Am. J. Surg. Pathol. 30, 828–837 (2006).
Bi, W. L. et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 69, 127–157 (2019).
van der Voort, S. R. et al. Predicting the 1p/19q Codeletion Status of Presumed Low-Grade Glioma with an Externally Validated Machine Learning Algorithm. Clin. Cancer Res. 25, 7455–7462 (2019).
Akkus, Z. et al. Predicting Deletion of Chromosomal Arms 1p/19q in Low-Grade Gliomas from MR Images Using Machine Intelligence. J. Digit Imaging. 30, 469–476 (2017).
Han, Y. et al. Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J. Neurooncol. 140, 297–306 (2018).
Kong, Z. et al. Thin-Slice Magnetic Resonance Imaging-Based Radiomics Signature Predicts Chromosomal 1p/19q Co-deletion Status in Grade II and III Gliomas. Front. Neurol. 11, 551771 (2020).
Afshar, P. et al. From handcrafted to deep-learning based cancer radiomics: challenges and opportunities. IEEE Signal Processing Magazine. 19, 132–160 (2019).
Truhn, D. et al. Radiomic versus Convolutional Neural Networks Analysis for Classification of Contrast-enhancing Lesions at Multiparametric Breast MRI. Radiology. 290, 290–297 (2019).
Sun, Q. et al. Deep Learning vs. Radiomics for Predicting Axillary Lymph Node Metastasis of Breast Cancer Using Ultrasound Images: Don’t Forget the Peritumoral Region. Front. Oncol. 10, 53 (2020).
Kermany, D. S. et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 172, 1122–1131.e9 (2018).
Yogananda, C. G. B. et al. A novel fully automated MRI-based deep-learning method for classification of 1p/19q co-deletion status in brain gliomas. Neurooncol. Adv. 2, vdaa066 (2020).
Zhang, Z. Y. et al. Prognostic value of preoperative hematological markers combined with molecular pathology in patients with diffuse gliomas. Aging 11, 6252–6272 (2019).
Cagney, D. N. et al. The FDA NIH Biomarkers, EndpointS, and other Tools (BEST) resource in neuro-oncology. Neuro. Oncol. 20, 1162–1172 (2018).
Wijnenga, M. M. J. et al. The impact of surgery in molecularly defined low-grade glioma: an integrated clinical, radiological, and molecular analysis. Neuro. Oncol. 20, 103–112 (2018).
Clark, V. E. & Cahill, D. P. Extent of resection versus molecular classification: what matters when. Neurosurg. Clin. N Am. 30, 95–101 (2019).
Ding, X. et al. The prognostic value of maximal surgical resection is attenuated in oligodendroglioma subgroups of adult diffuse glioma: a multicenter retrospective study. J. Neurooncol. 140, 591–603 (2018).
Kawaguchi, T. et al. Impact of gross total resection in patients with WHO grade III glioma harboring the IDH 1/2 mutation without the 1p/19q co-deletion. J. Neurooncol. 129, 505–514 (2016).
Patel, S. H. et al. T2-FLAIR Mismatch, an Imaging Biomarker for IDH and 1p/19q Status in Lower-grade Gliomas: A TCGA/TCIA Project. Clin. Cancer Res. 23, 6078–6085 (2017).
Goyal, A. et al. The T2-FLAIR-mismatch sign as an imaging biomarker for IDH and 1p/19q status in diffuse low-grade gliomas: a systematic review with a Bayesian approach to evaluation of diagnostic test performance. Neurosurg. Focus 47, E13 (2019).
Kim, M. et al. Diffusion- and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur. Radiol. 30, 2142–2151 (2020).
Choi, K. S., Choi, S. H. & Jeong, B. Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro. Oncol. 21, 1197–1209 (2019).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. U20A20171, 82102149, 61901458, 81702465, 61571432 and U1804172 and U1904148), the Science and Technology Program of Henan Province (No. 182102310113, 192102310123, and 192102310050), The Key Program of Medical Science and Technique Foundation of Henan Province (No. SBGJ202002062), The Joint Construction Program of Medical Science and Technique Foundation of Henan Province (No. LHGJ20190156), and Youth Innovation Promotion Association of the Chinese Academy of Sciences (2018364).
Author information
Authors and Affiliations
Contributions
Research conception: Z.Y.Z., J.Y., Z.-C.L., X.L., J.C., and W.L.; data processing, drafting of manuscript: Z.Y.Z., J.Y., Z.-C.L.i, S.Z., Wei.W., L.W., T.D., and Q.S.; data acquisition: D.P., W.D., C.S., Wen.W., Z.L., X.H., Y.G., and X.W.; data verification: D.P., W.D.; all authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was a part of the registered clinical trial “MR Based Prediction of Molecular Pathology in Glioma Using Artificial Intelligence” (ClinicalTrials.gov ID: NCT04217018). This study was approved by the Human Scientific Ethics Committee of the First Affiliated Hospital of Zhengzhou University (No. 2019-KY-176). Informed consents were waived by the Committee due to the retrospective and anonymous nature.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yan, J., Zhang, S., Sun, Q. et al. Predicting 1p/19q co-deletion status from magnetic resonance imaging using deep learning in adult-type diffuse lower-grade gliomas: a discovery and validation study. Lab Invest 102, 154–159 (2022). https://doi.org/10.1038/s41374-021-00692-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00692-5
This article is cited by
-
Artificial intelligence in neuro-oncology: advances and challenges in brain tumor diagnosis, prognosis, and precision treatment
npj Precision Oncology (2024)
-
Clinical and imaging characteristics of supratentorial glioma with IDH2 mutation
Neuroradiology (2024)
-
Two-headed UNetEfficientNets for parallel execution of segmentation and classification of brain tumors: incorporating postprocessing techniques with connected component labelling
Journal of Cancer Research and Clinical Oncology (2024)
-
Radiomic features from multiparametric magnetic resonance imaging predict molecular subgroups of pediatric low-grade gliomas
BMC Cancer (2023)
-
A novel MRI-based deep learning networks combined with attention mechanism for predicting CDKN2A/B homozygous deletion status in IDH-mutant astrocytoma
European Radiology (2023)