Abstract
Dysregulation of long noncoding RNA (LncRNA) FENDDR has been shown to be closely related to the progression of several cancers. However, its role and upstream regulatory mechanism in endometrioid endometrial carcinoma (EEC) remains unclear. This study was conducted using the cancerous tissues of EEC patients (n = 60), EEC cell lines, and a xenograft mouse model. The expression level of LncRNA FENDRR was decreased and the N-methyladenosine (m6A) methylation levels of LncRNA FENDRR was elevated in cancerous tissues of EEC patients. In vitro experiments demonstrated that YTH domain-containing 2 (YTHDF2), an m6A reader, recognized the abundance of m6A-modified LncRNA FENDRR in EEC cells and promoted its degradation. LncRNA FENDRR overexpression suppressed cell proliferation and facilitated cell apoptosis in the EEC cell line HEC-1B by reducing the protein level of SRY-related HMG box transcription factor 4 (SOX4). Interference of LncRNA FENDRR reversed the inhibitory effect of sh-YTHDF2 on cell proliferation and the promoting effect of sh-YTHDF2 on cell apoptosis in HEC-1B cells by silencing FENDRR. Finally, in vivo experiments confirmed that overexpression of LncRNA FENDRR retarded the growth of EEC cells. In conclusion, YTHDF2-mediated LncRNA FENDRR degradation promotes cell proliferation by elevating SOX4 expression in EEC.
Similar content being viewed by others
Introduction
Endometrial cancer (EC) is one of the most prevalent gynecologic malignancies, causing ~90,000 deaths worldwide in 2018 [1]. According to conventional histopathology, EC is divided into two pathological types: endometrioid endometrial carcinoma (EEC) and non-endometrioid endometrial carcinomas [2]. Among them, EEC is estrogen dependent and accounts for 80% of total cases [3]. Although the majority of EEC patients are diagnosed early with a favorable prognosis, 20% die within five years [4]. Therefore, gaining a better understanding of the mechanisms underlying EEC progression is of great importance, as it will help identify new therapeutic targets for EEC.
N6-methyladenosine (m6A) is the most abundant modification of eukaryotic RNA molecules, and it plays a vital role in processes such as RNA transcription, processing, and stability [5]. M6A modification is mainly regulated by three essential factors: m6A methyltransferases, demethylases, and m6A-binding proteins [6]. The methyltransferase complex that is composed of methyltransferase-like 3 (METTL3), METTL14, and Wilms tumor 1-associated protein, contributes to the initiation of m6A modification [7, 8]. M6A demethylases, such as obesity-associated protein (FTO) and alkB homolog 5, remove m6A from RNAs to dynamically regulate m6A modification [9, 10]. M6A-binding proteins (also known as m6A readers), including YTH domain-containing 2 (YTHDF2), specifically recognizes m6A modifications and are responsible for the regulatory effect of the m6A group on downstream RNAs [11]. Several studies have revealed that m6A-mediated RNA regulation modulates the pathological progression of various cancers [12, 13]. Importantly, Liu et al. [14] reported that m6A methylation was reduced in EC, leading to downregulation of negative AKT regulator PHLPP2 and upregulation of positive AKT regulator mTORC2. This, in turn, results in increased cell proliferation by activating the AKT pathway. However, the role of m6A modification in the development of EEC is still unknown.
Long noncoding RNA (LncRNA) FENDRR is a well-known tumor-suppressor gene in various cancers, such as breast cancer [15], gastric cancer [16], and prostate cancer [17]. Recently, LncRNA FENDRR has been identified as a dysfunctional molecule in estrogen receptor alpha-related EC [18]. Liu et al. [19] proved that LncRNA FENDRR suppressed the protein level of SRY-related HMG box transcription factor 4 (SOX4) in colon cancer cell lines. Given that the SOX4 level is elevated in tumor tissues of EEC patients and its knockdown was found to suppress cell proliferation in EEC cell lines [20], we speculated that LncRNA FENDRR may play a role in EEC progression by modulating SOX4 expression. In addition, utilizing a public prediction server called SRAMP, we identified seven m6A recognition sites in the sequence of LncRNA FENDRR, indicating that LncRNA FENDRR may be susceptible to m6A methylation modification.
In the present study, we investigate the functional role of LncRNA FENDRR in EEC progression and determine whether m6A modification participates in EEC progression by modulating LncRNA FENDRR.
Materials and methods
Tissue samples
Endometrial cancerous tissues were obtained from 60 EEC patients who underwent surgical excision at The First Affiliated Hospital, College of Medicine, Zhejiang University. All EEC patients were histologically validated for type and grade and did not undergo preoperative chemotherapy or radiation before surgery. Normal endometrial tissues (NE) were collected from 60 hysteromyoma patients who received hysterectomies. The study was approved by the Ethics Committee of The First Affiliated Hospital, Zhejiang University School of Medicine. Before the study began, all patients enrolled in the study signed informed consent forms.
Cell culture and transfection
Procell Life Science & Technology Co., Ltd. (China) provided the human EEC cell lines (Ishikawa and HEC-1B) used in this study. Ishikawa cells were cultured with Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum (FBS). HEC-1B cells were cultured with Eagle’s minimum essential medium containing 10% FBS.
RiboBio (China) synthesized pcDNA-FTO, pcDNA-FENDRR, pcDNA-SOX4, sh-YTHDF2, sh-FENDRR, and their negative controls (pcDNA and shRNA). For transient transfection, Ishikawa and HEC-1B cells were transfected with the indicated plasmids utilizing Lipofectamine 3000 reagent (Invitrogen, USA).
qRT-PCR
Total RNA samples were extracted from tissues and EEC cell lines using Trizol reagent (Sangon Biotech, China) and reversely transcribed into cDNA. Then, SYBR-Green PCR Master Mix (Applied Biosystems, CA) was used to conduct qRT-PCR. The relative expression levels of LncRNA FENDRR and SOX4 were calculated using the 2−ΔΔCT method. GAPDH served as an endogenous control. The primer sequences used for qRT-PCR were as follows: FENDRR 5′-AGACAAAAACTCACTGCCCA-3′ (F) and 5′-TGATGTTCTCCTTCTTGCCTC-3′ (R); SOX4 5′-ACAATGCCGAGAACACGGAAGC-3′(F) and 5′-GATCTGCGACCACACCATGAAGG-3′ (R); and GAPDH 5′-TGTTCGTCATGGGTGTGAAC-3′ (F) and 5′-ATGGCATGGACTGTGGTCAT-3′ (R).
Methylated RNA immunoprecipitation (MeRIP) assay
Total RNA samples were extracted from tissues and EEC cell lines, followed by incubation with an anti-m6A antibody. Then, the immunoprecipitate was reversely transcribed into cDNA, and the level of m6A-methylated LncRNA FENDRR was determined by qRT-PCR.
RNA immunoprecipitation (RIP)
The lysate of Ishikawa and HEC-1B cells was prepared. Anti-YTHDF2 or anti-IgG antibody was prebound to the protein A/G beads and then reacted with the cell lysate overnight at 4 °C. After elution from the beads, the RNA samples were precipitated with ethanol and dissolved in RNase-free water. The relative expression level of LncRNA FENDRR in the immunoprecipitate was determined by qRT-PCR.
RNA pull-down assay
A biotin-labeled LncRNA FENDRR probe was provided by RiboBio (China). Cell lysates of Ishikawa and HEC-1B cells were incubated with the biotin-labeled probe, followed by interaction with streptavidin-coupled magnetic beads. The expression level of LncRNA FENDRR in the complex pulled down by the biotin-labeled LncRNA FENDRR probe was measured using Western blot analysis.
Western blot
Western blot analysis was conducted to determine the protein levels of YTHDF2 and SOX4, with GAPDH serving as an internal control. Primary antibodies against YTHDF2 (ab220163) and GAPDH (ab9485) were purchased from Abcam (UK) and primary antibody against SOX4 (PA5-41442) was purchased from Thermo Fisher (USA).
Cell proliferation assay
A 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay was performed to evaluate the proliferation of HEC-1B cells. Before measurement, HEC-1B cells were transfected with plasmids and seeded into 96-well plates. Cells were incubated with MTS (20 ul) at 37 °C at five-time points after transfection (0, 24, 48, 72, and 96 h). One hour later, the absorbance of each well at 490 nm was measured using a microplate reader.
Cell apoptosis assay
The apoptosis of HEC-1B cells was measured using flow cytometry as previously described [21]. HEC-1B cells were transfected with plasmids and seeded into 6-well plates. For measurement, HEC-1B cells were resuspended in the binding buffer and then orderly incubated with Annexin V-FITC for 15 min and Propidium Iodide Staining Solution for 5 min. Cell apoptosis was measured by a flow cytometer (BD Biosciences, USA).
Xenograft growth assay
The antitumor effect of LncRNA FENDRR in EEC was evaluated by the xenograft model. A total of 12 BALB/C nude mice (female, six-week-old) were purchased from Beijing Vital River Laboratory Animal Technologies Co. Ltd (China). HEC-1B cells (1 × 107) transfected with pcDNA or pcDNA-FENDRR were subcutaneously transplanted into the backs of the mice. Three weeks after injection, the tumor volume of mice in the pcDNA (n = 6) and pcDNA-FENDRR (n = 6) groups was measured at one-week intervals. Eight weeks after injection, all mice were sacrificed, and the tumor tissues were collected. The Ethics Committee of The First Affiliated Hospital, Zhejiang University School of Medicine approved all animal experiments conducted as part of this study.
Statistical analysis
In vitro data were obtained from three replicates of three independent experiments. Statistical analysis was conducted using SPSS 19.0. Data were expressed as mean ± SD and compared (between two experimental groups) utilizing a Student’s t test. The results were deemed statistically significant when P < 0.05.
Results
LncRNA FENDRR expression was downregulated in the cancerous tissues of EEC patients
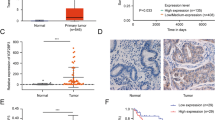
The results of qRT-PCR depicted that the LncRNA FENDRR expression level was decreased in EEC cancerous tissues compared with NE (Fig. 1A). The lower expression of LncRNA FENDRR was statistically correlated with more advanced histologic grade, lymph node metastasis, and recurrence of EEC patients (Table 1), indicating the potential of LncRNA FENDRR to function as a biomarker of the malignant progression of EEC. The expression level of SOX4, measured by Western blot, indicated that SOX4 protein level was increased in the EEC cancerous tissues compared with NE (Fig. 1B). In addition, a public prediction server (SRAMP) forecasted seven m6A recognition sites in the sequence of LncRNA FENDRR (Fig. 1D). Through performing the MeRIP-qPCR assay, we verified that the m6A modification was indeed enriched in LncRNA FENDRR. What’s more, compared with NE, the level of m6A-modified LncRNA FENDRR was significantly increased in EEC cancerous tissues (Fig. 1C).
The expression levels of (A) LncRNA FENDRR and (B) SRY-related HMG box transcription factor 4 (SOX4) were measured by qRT-PCR and Western blot in EEC cancerous tissues (n = 60) and normal endometrial tissues (NE; n = 60), respectively. ***P < 0.001. C Methylated RNA immunoprecipitation (MeRIP) assay was performed using an anti-m6A antibody in EEC cancerous tissues and NE. Enrichment of m6A-modified LncRNA FENDRR was then analyzed through qRT-PCR. *P < 0.05 vs NE. D The potential m6A sites in the sequence of LncRNA FENDRR predicted by SRAMP.
Degradation of m6A-methylated FENDRR was promoted by YTHDF2 in EEC cell lines
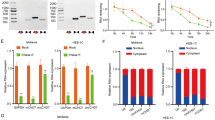
To further explore the influence of m6A modification on LncRNA FENDRR expression, FTO, a type of m6A demethylase, was overexpressed in EEC cell lines through pcDNA-FTO transfection. As shown in Fig. 2A, m6A enrichment in LncRNA FENDRR was notably decreased upon pcDNA-FTO transfection, indicating that transfection successfully overexpressed FTO and reduced m6A modification in both Ishikawa and HEC-1B cells. Interestingly, LncRNA FENDRR expression was prominently elevated in response to FTO overexpression (Fig. 2B), suggesting that the expression level of LncRNA FENDRR was negatively regulated by m6A modification. A previous study demonstrated that m6A reader YTHDF2 directly bound m6A-modified LncRNA GAS5 and decreased its stability [22]. So we aimed to determine whether m6A modification reduced LncRNA FENDRR expression by recruiting YTHDF2. A RIP assay revealed a greater accumulation of LncRNA FENDRR in YTHDF2 immunoprecipitate compared with IgG immunoprecipitate (Fig. 2C). The results of the RNA pull-down showed that an abundance of YTHDF2 protein was presented in the complex pulled down by LncRNA FENDRR (Fig. 2D). These data confirmed that LncRNA FENDRR was specially recognized by YTHDF2. Then, the effect of YTHDF2 on the stability of LncRNA FENDRR was measured in the presence of actinomycin D, an inhibitor of RNA synthesis. As depicted in Fig. 2E, YTHDF2 knockdown efficiently decreased the degradation of LncRNA FENDRR.
The EEC cell lines Ishikawa and HEC-1B were transfected with pcDNA-obesity-associated protein (FTO) or its negative control (pcDNA). A MeRIP-qPCR assay was performed to measure the enrichment of m6A-modified LncRNA FENDRR. *P < 0.05 vs pcDNA. B The LncRNA FENDRR expression was measured by qRT-PCR. *P < 0.05 vs pcDNA. C RNA-binding protein immunoprecipitation (RIP) followed by qRT-PCR was performed to measure the endogenous combination of LncRNA FENDRR and YTH domain-containing 2 (YTHDF2). *P < 0.05 vs IgG. D RNA pull-down assay verified LncRNA FENDRR was specifically recognized by YTHDF2. EGFP RNA was used as an RNA control. E Ishikawa and HEC-1B cells were transfected with sh-YTHDF2 or its negative control (shRNA), followed by actinomycin D (5 μg/ml) treatment. Upper: LncRNA FENDRR expression level was determined using qRT-PCR at indicated times. *P < 0.05 vs shRNA. Under: the transfection efficiency of sh-YTHDF2 was confirmed by Western blot.
LncRNA FENDRR overexpression suppressed cell proliferation through decreasing SOX4 expression in EEC cell lines
As reported, the interference of SOX4 could repress the proliferation of EEC cell lines [20]. Meanwhile, the expression level of SOX4 was negatively regulated by LncRNA FENDRR in colon cancer cells [19]. To investigate whether LncRNA FENDRR took part in the progression of EEC by regulating SOX4 expression, the following experiments were conducted. First, the mRNA and protein levels of SOX4 were determined in EEC cell lines transfected with pcDNA or pcDNA-FENDRR. We found that the protein level of SOX4, rather than the mRNA level, was notably decreased after LncRNA FENDRR overexpression (Fig. 3A, B). Second, HEC-1B cells were transfected with pcDNA, pcDNA-FENDRR, or pcDNA-FENDRR + pcDNA-SOX4. The results depicted in Fig. 3C, D confirmed that pcDNA-FENDRR transfection efficiently upregulated LncRNA FENDRR level and pcDNA-SOX4 transfection rescued the protein level of SOX4 reduced by LncRNA FENDRR overexpression. Moreover, in response to LncRNA FENDRR overexpression, cell proliferation was decreased and cell apoptosis was elevated in HEC-1B cells (Fig. 3E, F). On the contrary, in HEC-1B cells cotransfected with pcDNA-SOX4 + pcDNA-FENDRR, pcDNA-SOX4 increased cell proliferation and suppressed cell apoptosis in the presence of pcDNA-FENDRR (Fig. 3E, F). This indicates that SOX4 mediated the modulatory effect of LncRNA FENDRR on cell proliferation and apoptosis in EEC cells.
The (A) mRNA and (B) protein levels of SRY-related HMG box transcription factor 4 (SOX4) were measured in EEC cell lines transfected with pcDNA-FENDRR or its negative control (pcDNA). GAPDH was used as an internal control. HEC-1B cells were divided into control, pcDNA, pcDNA-FENDRR, and pcDNA-FENDRR + pcDNA-SOX4 groups. C LncRNA FENDRR expression was determined using qRT-PCR. D The mRNA and protein levels of SOX4 were determined by qRT-PCR and Western blot, respectively. E Cell proliferation was determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay. F Cell apoptosis was determined using flow cytometry. *P < 0.05.
YTHDF2 knockdown suppressed proliferation of EEC cells by reducing m6A-modified LncRNA FENDRR degradation
Since we confirmed that YTHDF2 knockdown reduced the degradation of m6A-modified LncRNA FENDRR (Fig. 2) and LncRNA FENDRR overexpression suppressed the proliferation of EEC cells (Fig. 3), we explored whether YTHDF2 knockdown could repress cell proliferation by elevating LncRNA FENDRR expression. As shown in Fig. 4A, sh-YTHDF2 significantly elevated LncRNA FENDRR expression, while the promotion effect of sh-YTHDF2 on LncRNA FENDRR expression was abrogated by sh-FENDRR. Similarly, sh-FENDRR reversed the inhibitory effect of sh-YTHDF2 on the protein level of SOX4 (Fig. 4B). After sh-YTHDF2 transfection, cell proliferation was downregulated, while cell apoptosis was upregulated (Fig. 4C, D). However, sh-FENDRR elevated cell proliferation and reduced cell apoptosis in the presence of sh-YTHDF2 (Fig. 4C, D). As shown in Fig. 4E, LncRNA FENDRR became m6A methylated after pcDNA-FENDRR transfection and the methylated-FENDRR level peaked 36 h after transfection. However, the expression level of LncRNA FENDRR was not significantly decreased because of abundant pcDNA-FENDRR transfection (Fig. 4F). These results indicated that m6A methylation is not sufficient to change the level of FENDRR with pcDNA-FENDRR transfection. Hence, the expression of FENDRR remained high after demethylation with FTO transfection, and demethylation had a limited impact on cell proliferation (Fig. 4G) and apoptosis (Fig. 4H).
(A–D) HEC-1B cells were divided into control, shRNA, sh-YTHDF2, and sh-YTHDF2 + sh-FENDRR groups. A LncRNA FENDRR expression was determined using qRT-PCR. B The protein levels of SOX4 were determined by Western blot. C Cell proliferation was determined by MTS assay. D Cell apoptosis was determined using flow cytometry. (E–H) HEC-1B cells were divided into pcDNA, pcDNA-FENDRR, and pcDNA-FENDRR + pcDNA-FTO groups. E MeRIP-qPCR assay was performed to measure the enrichment of m6A-modified LncRNA FENDRR. F LncRNA FENDRR expression was determined using qRT-PCR. G Cell proliferation was determined by MTS assay. H Cell apoptosis was determined using flow cytometry. *P < 0.05.
LncRNA FENDRR overexpression retarded the growth of EEC cells in vivo
In order to verify the influence of LncRNA FENDRR on tumorigenesis in vivo, HEC-1B cells transfected with pcDNA or pcDNA-FENDRR were subcutaneously injected into mice. By detecting the expression level of LncRNA FENDRR in the tumor tissues of mice, we confirmed that LncRNA FENDRR was successfully overexpressed in mice transplanted with pcDNA-FENDRR-transfected HEC-1B cells (Fig. 5C). Compared with mice transplanted with pcDNA-transfected HEC-1B cells, the mice transplanted with pcDNA-FENDRR-transfected HEC-1B cells exhibited smaller tumor volume and lower tumor weight (Fig. 5A, B). Besides, similar to in vitro experiments, LncRNA FENDRR overexpression reduced the SOX4 protein level (Fig. 5D). The above data hinted that LncRNA FENDRR exerted its antitumor effect by suppressing SOX4 expression in the xenograft mouse model.
1 × 107 HEC-1B cells were transfected with pcDNA or pcDNA-FENDRR, followed by subcutaneously injected into the nude mice. n = 6 of each group. Mice were sacrificed eight weeks after injection. A The tumor volume of mice was measured at the indicated times. B The tumor weight of mice in each group. C LncRNA FENDRR expression in the tumor tissues of mice was determined using qRT-PCR. D The protein level of SOX4 in the tumor tissues of mice was determined using Western blot. *P < 0.05 vs pcDNA.
Discussion
Increasing evidence reveals that LncRNAs, a class of ncRNAs >200 nucleotides in length, are closely related to a series of biological processes during EEC progression [23, 24]. In this study, we found that the m6A modification level of LncRNA FENDRR was elevated in EEC patients, and abundant m6A modification promoted LncRNA FENDRR degradation by recruiting the m6A binding protein YTHDF2. Subsequently, downregulation of LncRNA FENDRR resulted in the accumulation of SOX4 protein, thus boosting the proliferation of EEC cells and further aggravating the pathological process of EEC (Fig. 6).
As the most abundant modification on eukaryotic RNA molecules, the role of m6A methylation in cancer has attracted widespread interest among researchers. Lin et al. [25] found that METTL3, an m6A methyltransferase, boosted the growth of human cancer cells by facilitating the translation of oncogenes. Zhong et al. [26] reported that YTHDF2 destabilized the mRNA of epidermal growth factor receptor by binding the m6A site in its 3′UTR of mRNA, thus suppressing the proliferation of hepatocellular carcinoma cells. In addition to mRNA, the stability of LncRNAs is modulated by YTHDF2 during cancer progression. The research of Wang et al. [22] proved that in cervical cancer cells, YTHDF2 negatively regulated the expression of LncRNA GAS5 by facilitating its degradation. Consistent with their study, we found that overexpression of m6A demethylase FTO increased the level of LncRNA FENDRR, and knockdown of YTHDF2 increased the level of LncRNA FENDRR by suppressing its degradation. These results prove that the promotion effect of m6A modification on LncRNA FENDRR level is dependent on YTHDF2. Then, the following experiments show that the interference of YTHDF2 significantly decreased proliferation and promoted apoptosis of EEC cells by restoring LncRNA FENDRR expression, suggesting the potential tumor-inducing function of YTHDF2 in EEC.
Researchers have verified the antitumor effect of LncRNA FENDRR in various cancers [15,16,17], but to date, the role of LncRNA FENDRR in EEC has remained unclear. In our study, the low expression level of LncRNA FENDRR in the tumor tissues of EEC patients and the close correlation between low expression of LncRNA FENDRR and malignant progression of EEC suggested the potential role of LncRNA FENDRR in EEC. As speculated, our in vitro experiments showed that overexpression of LncRNA FENDRR effectively suppressed cell proliferation and induced cell apoptosis in EEC cell lines by downregulating SOX4 protein level. These results are supported by Liu et al. [19], who found a negative regulatory effect of LncRNA FENDRR on SOX4 expression in colon cancer. Considering that LncRNAs can modulate the expression level of the target protein by influencing its stability [27], in our follow-up research, we will further explore the specific regulatory mechanism of LncRNA FENDRR on SOX4.
To sum up, our study revealed the vital role of m6A modification in the downregulation of LncRNA FENDRR in EEC, and it provided evidence that LncRNA FENDRR could function as a tumor suppresser gene in EEC by suppressing cell proliferation.
References
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–53.
Makker A, Goel MM. Tumor progression, metastasis, and modulators of epithelial-mesenchymal transition in endometrioid endometrial carcinoma: an update. Endocr Relat Cancer. 2016;23:R85–111.
Geels YP, van der Putten LJ, van Tilborg AA, Lurkin I, Zwarthoff EC, Pijnenborg JM, et al. Immunohistochemical and genetic profiles of endometrioid endometrial carcinoma arising from atrophic endometrium. J Gynecol Oncol. 2015;137:245–51.
Guan J, Xie L, Luo X, Yang B, Zhang H, Zhu Q, et al. The prognostic significance of estrogen and progesterone receptors in grade I and II endometrioid endometrial adenocarcinoma: hormone receptors in risk stratification. J Gynecol Oncol. 2019;30:e13.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Zhao W, Qi X, Liu L, Ma S, Liu J, Wu J. Epigenetic regulation of m(6)A modifications in human cancer. Mol Ther Nucleic Acids. 2020;19:405–12.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83.
Li Y, Zhang W, Liu P, Xu Y, Tang L, Chen W, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. 2018;11:1403–12.
Li L, Yang WT, Zheng PS, Liu XF. SOX17 restrains proliferation and tumor formation by down-regulating activity of the Wnt/beta-catenin signaling pathway via trans-suppressing beta-catenin in cervical cancer. Cell Death Dis. 2018;9:741.
Zhang YQ, Chen X, Fu CL, Zhang W, Zhang DL, Pang C, et al. FENDRR reduces tumor invasiveness in prostate cancer PC-3 cells by targeting CSNK1E. Eur Rev Med Pharmacol Sci. 2019;23:7327–37.
Liu A, Zhang D, Yang X, Song Y. Estrogen receptor alpha activates MAPK signaling pathway to promote the development of endometrial cancer. J Cell Biochem. 2019;120:17593–601.
Liu J, Du W. LncRNA FENDRR attenuates colon cancer progression by repression of SOX4 protein. Onco Targets Ther. 2019;12:4287–95.
Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–46.
Zhou YX, Mao LW, Wang YL, Xia LQ, Zhao W. Increased LncRNA PVT-1 is associated with tumor proliferation and predicts poor prognosis in cervical cancer. Clin Surg Res Commun. 2017;1:10–17.
Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–21.
Yang X, Wang CC, Lee WYW, Trovik J, Chung TKH, Kwong J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett. 2018;413:23–34.
Li Z, Wei D, Yang C, Sun H, Lu T, Kuang D. Overexpression of long noncoding RNA, NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother. 2016;84:244–51.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62:335–45.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–61.
Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73–80.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Shen, J., Feng, Xp., Hu, Rb. et al. N-methyladenosine reader YTHDF2-mediated long noncoding RNA FENDRR degradation promotes cell proliferation in endometrioid endometrial carcinoma. Lab Invest 101, 775–784 (2021). https://doi.org/10.1038/s41374-021-00543-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00543-3
This article is cited by
-
Novel insights into the multifaceted roles of m6A-modified LncRNAs in cancers: biological functions and therapeutic applications
Biomarker Research (2023)
-
A methylation- and immune-related lncRNA signature to predict ovarian cancer outcome and uncover mechanisms of chemoresistance
Journal of Ovarian Research (2023)
-
Novel insights into mutual regulation between N6-methyladenosine modification and LncRNAs in tumors
Cancer Cell International (2023)
-
METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling
Cell Death & Disease (2023)
-
The roles of N6-methyladenosine and its target regulatory noncoding RNAs in tumors: classification, mechanisms, and potential therapeutic implications
Experimental & Molecular Medicine (2023)