Abstract
Cellular senecence is an important biologic endpoint. Naturally occuring (aging) senescence is common in uterine leiomyoma (ULM). AKT is one of major pathways in promoting ULM growth and survival. Inactivation of AKT by MK2206 in ULM resulted in stress-induced senescence in vitro. Study of the senescent phenotypes and molecular changes in ULM may greatly facilitate the understanding of the tumor biology and potential clinical therapy for this common disease associated with high morbidity. To study senescence in a model system that closely resembles primary ULM in vivo, we applied an ex vivo model of three-dimensional (3D) spheroid culture system which maintained the molecular and cellular characteristics of primary ULM and matched myometrium as seen in vivo. Gene expression profiling done on ULM induced to undergo replication (passaging) or stress-induced (MK2206) senescence revealed that ROS and hypoxic-related genes were upregulated in the two types of senescences. Overexpression of two selected genes, WIPI1 and SLITKR4, induced cellular senescence in ULM spheroids. Additionally, administration of ABT263 (a BH3 mimetic) effectively reduced the senescent cells induced in ULM spheroids. This study identified novel genes associated with senescence in ULM and demonstrated a BH3 mimetic to act as a senolytic to remove senescent cells.
Similar content being viewed by others
Introduction
Uterine leiomyomas (ULM) are benign uterine smooth muscle tumors and are the most common neoplasms in women and up to 70% of women develop ULM during their reproductive ages [1]. ULM cause significant morbidity due to profuse menstrual bleeding, pelvic discomfort, as well as reproductive dysfunction [2]. The current treatment options include cytoreductive surgeries, such as myomectomy and hysterectomy, and anti-hormonal treatments, such as birth control pills and gonadotropin-releasing hormone (GnRH) analogs, which can only temporarily reduce the tumor size but induce side effects [3]. Therefore, non-hormonal, non-invasive therapeutic modalities are attractive alternatives.
The pathophysiology and growth control in ULM are not well characterized. According to clinical observations, the rate of fibroid growth varies widely in women of different ages, hormonal status, tumor microenvironment, and the specific gene mutations that drive the tumor [4]. Our previous study showed that about half of ULM exhibited focal and patchy senescence [5]. The causes of cellular senescence in ULM are largely unknown and may be related to aging or local stress [6, 7]. Studies show that ULM are in a hypoxic microenvironment and have high ROS exposure due to a defective ROS metabolic pathway [8]. The AKT pathway is a major growth and survival pathway for ULM under oxidative stress. Treating leiomyoma cells in vitro with the AKT allosteric inhibitor can trigger stress-induced premature senescence in leiomyoma cells [7]. These findings provide a promising venue to develop a new therapeutic approach in controlling ULM growth. Furthermore, the recent development of anti-BCL2 molecules which can effectively destroy most senescent cells (as senolytic molecules) provide an improved solution that avoids the possible side effect of many inflammatory cytokines produced by senescent cells. For these reasons, we set out to investigate the senescence phenotype, molecular mechanisms and potential therapeutic strategy in a novel model that closely mimics ULM in vivo.
In this study, we investigated the senescence phenotypes of naturally occurring replication or stress induced senescence in primary ULM. An ex-vivo three-dimensional culture system (spheroids) were used as they maintained certain cellular and molecular characteristics of primary ULM. Gene expression profiling on both replicative or AKT-inhibitor induced cellular senescence in primary fibroid cells identified novel senescence associated genes, two of which were further investigated for its ability to induce senescence. Finally the senolytic effect of ABT263 on senescent cells in ULM spheroids was evaluated.
Materials and methods
Collection of tissue samples and culture of primary cells
Human myometrium and leiomyoma tissue samples were collected from premenopausal women undergoing hysterectomy at the Northwestern University Prentice Women’s Hospital (Chicago, Illinois). All the patients that participated in this study that were not taking hormonal contraceptives for at least 3 months at the time of surgery were consented (according to the IRB (International Review Board) approved protocol). The patients’ age, ethnicity, hormonal status, reason for surgery, MED12 mutation, HMGA2 translocation, and FH alteration are summarized in supplementary Table 1. To ensure the results were comparable and stable, only MED12 mutant ULM were used for this study. Among 28 hysterectomies for ULM, 43% (12/28) harbor MED12 mutations, 11% gain HMGA2 overexpression and no biallelic loss of FH is found. Primary myometrial and leiomyoma cells were digested and cultured as previously described [8]. Primary cells in monolayer were cultured either in Dulbecco’s modified Eagle’s medium/nutrient Ham’s Mixture F-12 (DMEM-F12) 1:1 containing 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin at 37 °C and 5% CO2 atmosphere for experiments that could be conducted within 5 days, or Smooth Muscle Growth Medium-2 (SmGMTM-2 medium) (Lonza) for serial passaging and long-term cultures (preventing from loss of smooth muscle cells in culture).
Senescence-associated β-galactosidase (SA-β-gal) staining
SA-β-gal staining was conducted following the manufacturer’s protocol (Cell Signaling Technology). Briefly, the cells were either passaged twice (P2) or treated with MK2206 at 5 μM (Selleck Chemicals). Cells were fixed with 2% formaldehyde plus 0.2% glutaraldehyde and stained with β-galactosidase staining solution (citric acid/sodium phosphate solution, potassium ferrocyanide, potassium ferricyanide, X-gal, pH 6) and incubated at 37 °C overnight in a dry incubator. The reactions were terminated when the cells were stained blue-green, as visualized under an inverted bright-field microscope. The cells were also stained with DAPI (4′, 6-Diamidino-2-Phenylindole, Dihydrochloride) to visualize the nucleus. The images were taken under the ×10 objective lens in three different fields (two-dimensional monolayer = 2D) or entire spheroid (three-dimensional nodule = 3D), and the percentage of cells (2D) and intensity of green color (3D) that were positive for β-galactosidase was calculated and averaged.
SDS-PAGE and western blotting
Protein lysates were extracted from myometrial and leiomyoma cells using RIPA lysis and extraction buffer with protease and phosphatase inhibitors (Thermo Fisher Scientific), and the protein concentration was determined using BCA Protein Assay kit (Thermo Fisher Scientific). Equal amounts of proteins were subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were then either blocked with 5% BSA in TBST or 5% milk in TBST. Immunoblotting was performed using the primary antibodies were summarized in Supplementary Table 2. Secondary antibodies were horseradish peroxidase (HRP)-labeled anti-mouse (7076S, Cell Signaling Technology) or anti-rabbit (7074S, Cell Signaling Technology) or anti-goat (HAF109, Fisher Scientific). Chemiluminescence was detected by adding a chemiluminescent HRP substrate (Thermo Fisher Scientific) and measured with a Fujifilm LAS-3000 Imager.
Gene expression microarray
Total RNA was isolated using the Rneasy Mini Kit (Qiagen) according to the manufacturer’s instructions. After elution, RNA quantity was assessed by NanoDrop 1000 spectrophotometer (NanoDrop Wilmington, DE). A microarray using the Illumina platform was performed with RNA samples that were either serial passaged or treated with MK2206 from four different patients. All RNA samples were processed at the Genomics Core Facility in the Center for Genetic Medicine at Northwestern University (Chicago, IL). Gene expression analysis was performed using the Illumina HumanHT-12 v4 Expression BeadChip. Expression data were normalized using the quartile normalization. After quality control and normalization, differentially expressed genes were identified through Volcano Plot Filtering.
RNA isolation and RT-PCR
RNA was isolated from uterine fibroid cells using Rneasy Mini Kit (Qiagen) and reverse-transcribed with M-MLV Reverse Transcriptase (Clontech) following the manufacturer’s instructions. Quantitative RT-PCR was performed using PowerUp™ SYBR® Green Master Mix (Life Technologies) on an Applied Biosystems® Real-Time PCR Instrument. Gene primers are summarized in Supplementary Table 3. GAPDH was used as the housekeeping gene, and relative mRNA levels were calculated using the 2−ΔΔCt method. Each data point is the average of three replicates.
cDNA transfection
WIPI1 (GFP-tagged)-cDNA (RG208094, OriGene Technology), SLITRK4 (GFP-tagged)-cDNA (RG207255, OriGene Technology) and pVector (GFP-tagged) control vector (PS100010, OriGene Technology) were transiently transfected into primary cells at 60% confluence using lipofectamine® 3000 transfection reagent (Thermo Fisher Scientific). After 3 days of transfection, cells were either harvested for western blot analysis or stained with β-galactosidase.
Spheroid cultures
Six-well plates were coated with 0.5% agarose gel which was allowed to solidify and cooled at room temperature (up to 3 mm spheroids in diameter) and/or 96-well black/clear bottom ultra-low attachment plates (Corning, up to 1 mm spheroids in diameter). Primary cells were resuspended and plated on top of the agarose, cultured with mesenchymal stem cell medium (Lonza, cat#190632 or cat#PT-3001). Cells were incubated in the humidified incubator with 5% CO2 at 37 °C for at least 48 h for formation of spheroids. For histology immunohistochemistry evaluation, spheroids were fixed in 4% paraformaldehyde (sc-281692, Santa Cruz Biotechnology) for 15 min, then switched to 100% alcohol overnight at 4 °C. Next day, spheroids were pelleted and placed in 0.5% agarose gel. The spheroids were processed, paraffin embedded, and sectioned. For β-galactosidase staining, spheroids were fixed with 2% formaldehyde plus 0.2% glutaraldehyde for 15 min and transferred to β-galactosidase staining solution listed below. For the cDNA transfection experiment, primary cells were transiently transfected as described above in 2D and then followed by the formation of spheroids.
Immunohistochemistry (IHC) staining of spheroids
Spheroids were fixed in 4% paraformaldehyde (sc-281692, Santa Cruz Biotechnology) for 15 min, then transferred to 100% alcohol overnight at 4 °C. The next day, spheroids were pelleted and placed in 0.5% agarose gel. The spheroids were fixed overnight, proceeded for tissue processing, embedded in paraffin, and sectioned. The H&E staining and immunostaining were performed by the Robert H. Lurie Comprehensive Cancer Center’s Pathology Core Facility. Immunostaining for pAKT, p16, p21, smooth muscle actin, Ki67, estrogen receptor (ER), progesterone receptor (PR) were performed as previously described [9] (Suppl Table 2).
Drug treatments
Two days after cell seeding, 50% of medium was replaced with fresh medium leaving the spheroids undisturbed. After two additional days, spheroids were treated with MK2206 (2–5 μM, Selleck Chemicals), ABT-263 (2 μM, AbbVie Inc, North Chicago, IL) or paraquat (PQ, Sigma-Aldrich) for 24–72 h at the indicated concentrations. Control spheroids were treated with an equivalent amount of the vehicle, DMSO.
Statistical analysis
GraphPad Prism was used for statistical analysis. All data were presented as means ± standard deviations. Student’s t-test was used to determine significance. P < 0.05 was considered statistically significant. Data from each patient were considered as an independent experiment.
Results
Analysis of three dimensional ULM spheroids
In vitro study for cellular senescence in monolayer culture has its limitation. To develop a culture system mimic to in vivo condition, we introduced ex vivo three-dimensional (3D) spheroid cultures. ULM cells, cultured in mesenchymal stem cell medium in 96-well plate, formed aggregates of cells that eventually took on a spheroid shape after 48–72 h (Fig. 1A1). The size of spheroids can be controlled depending on the initial number of loaded cells and the best size ranges were 0.5–3.0 mm in diameter. Monolayer culture can be restored from spheroids when transferred to routine culture dish and medium (Fig. 1A2). To evaluate the histology, the formalin-fixed and paraffin-embedded spheroids were prepared (see Materials and Methods). Histologic analysis revealed that ULM spheroids resembled the human primary myometrium (Fig. 1B1) and ULM (Fig. 1B2) architecture, characterized by thin smooth muscle bundles in myometrium, short fascicles and a storiform growth pattern in ULM. In particular, even after 5 days in culture, the thin and delicate newly synthesized collagen formation can be appreciated in Trichrome stain (Fig. 1C1). All observed histologic features can mimic to primary ULM seen in uterus (Fig. 1C2).
Three-dimensional spheroid culture for ULM cells. 3D spheroid nodule formation in 3D suspension culture illustrated in an invert light microscope (A1) and 3D spheroid nodule can reattach to form monolayer after transferring to regular culture plate (A2). Section of 3D spheroid nodules from myometrial (B1) and ULM (B2) on hematoxylin and eosin stained slides. Trichrome stain shows thin and delicate collagen (C1). A small primary ULM (C2) from uterus was presented as a comparison. Black bars indicated amplification
To further evaluate ULM-specific biomarkers as we commonly did in primary ULM, we conducted immunohistochemistry analysis of spheroids. After H&E staining, well-organized smooth muscle bundles in myometrial spheroids and storiform and a whorled appearance in ULM speroids could be appreciated (Figs. 1, 2). Spheroids revealed strong and diffusely immunoreactivity for smooth muscle actin (SMA) indicating that the majority of the cells in spheroids were smooth muscle cells. Estrogen and progesterone receptors (ER and PR) were also strongly positive by immunostaining (Fig. 2). Upon treatment of the ULM spheroids with 5μM MK2206 (AKT inhibitor, which were used extensive in our following assays), no significant change of ER and PR expression were observed (Fig. 2). The findings indicate that primary ULM spheroid cultures maintain the major histologic and molecular characters of normal myometrium and ULM.
Biomarker analysis of spheroids ULM. The immunohistochemistry analysis of formalin-fixed and paraffin-embedded myometrial (A) and leiomyoma (B) spheroids. Serial sections of spheroids from control (DMSO, left column) or test (MK2206, right column) myometrial and leiomyoma spheroids were examined and illustrated in routine H&E and immunohistochemistry stain for estrogen receptor (ER), progesterone receptor (PR) and smooth muscle actin (SMA) (10×)
Molecular analysis of replicative senescence in primary ULM cells
Replicative senescence can usually be observed when mitotically competent cells cease to proliferate permanently as a consequence of serial passaging of primary cells in culture [10]. To identify the genes that were differentially expressed in replication senescence in ULM, primary culture ULM cells were prepared from original cultures (P0) to two passages (P2). Since P2 were prepared in monolayer culture, to prevent from loss of smooth muscle cells, smooth muscle culture medium (SmGm2, see Methods) was used. In such condition, >80% of smooth muscle cells from myometrium and ULM can be preserved in long term culture (Suppl Fig. 1). ULM cells (at P0) from four patients were maintained in smooth muscle medium in primary culture and passaged twice (P2) until the cells ceased to grow and reached senescence as detected by SA-β-gal staining (Fig. 3a). Less than 20% of ULM cells in P0 were positive for SA-β-gal staining (Fig. 3a). After two passages, a significant increase of replicative senescence was found in ULM cells with 80% of cells staining positive for SA-β-gal staining in monolayer culture (p < 0.001). A similar global SA-β-gal staining pattern was observed in P0 and P2 spheroid ULM cells (Fig. 3a). To confirm the senescent phenotype, expression of p21 was examined in P0 and P2 cells (Fig. 3b). A significant increase of p16 and p21 expression was detected by western blot in primary leiomyomas from four patient-derived cells (Fig. 3b, Supplementary Figure 2).
Molecular and cellular analysis of replication senescence (serial passage) in primary ULM cells. a Cellular senescence was examined by SA-β-gal stain in the fresh prepared (P0) ULM cells and the ULM cells with two passages (P2). Left panels are monolayer (2D) and right spheroid (3D) culture ULM. Dark blue (2D) green (3D) stain is indicative of senescent cells. b Western blot analysis of p16 and p21 in P0 and P2 ULM cells. β-actin was used as a loading control. Relative expression levels were quantified by densitometry and illustrated by histobars (right, n = 4). c Differentially expressed genes (listed below) in P2 versus P0 were plotted in a heatmap and Dendragram. Color scale bar (right lower corner) indicates the relative expression levels from high (red) to low (blue). d Heatmap illustrates the upregulated genes highly enriched in ROS, hypoxia and DNA damage response pathway detected by pathway analysis in replication senescence. e, f Plot analysis of upregulated (e) and downregulated (f) genes in several functional pathways detected by GO pathway analysis
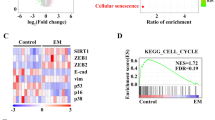
Next, gene expression profiling was performed using total RNA extracted from ULM cells at P0 and P2, as described above. After log2 conversion, normalization, quality control, and a fold change cutoff of 1.5, a total of 3243 genes were differentially expressed between P0 and P2 with statistical significance (p < 0.05, Fig. 3c). Among them, 1467 genes were upregulated and 1776 genes were downregulated. Pathway analysis by GO term showed that 211 upregulated (Fig. 3d) were enriched in several major functional pathways, involving in upregulation of ROS/hypoxia, DNA damage and AKT pathways (Fig. 3e). Of note, many genes in the p53 pathway and estrogen response were downregulated in replication senescence (Fig. 3f). This is consistent with recent findings that downregulation of p53 are essential for the induction of senescence-associated secretory phenotype (SASP) [11, 12]. The highly dysregulated genes (>1.5-fold change) in six pathways in replication senescence were listed in Suppl Table 4.
Molecular analysis in stress-induced senescence by AKT inhibitor MK2206 in primary ULM cells
MK2206, an allosteric AKT inhibitor, can effectively inhibit AKT as well as induce senescence [7]. ULM spheroids were treated with control (DMSO) and MK2206 (5 μM) for 24 h and examined by immunohistochemistry. Loss of immunoreactivity for pAKT was observed in MK2206 treated spheroids, but not control treated spheroids (Fig. 4a). After confirming MK2206 induced cellular senescence detected by SA-β-gal staining in both 2D and 3D ULM (Fig. 4b), p16 and p21expressions were demonstrated to be increased by immunohistochemistry (Fig. 4c) and western blot (Fig. 4d, Suppl Fig. 3). To evaluate gene expression changes in response to MK2206, microarray was performed using ULM cells from 4 cases treated with DMSO or MK2206. Overall the gene expression varied widely among different tumors in the MK2206-treated group, yielding less differentially expressed genes when compared with controls. After normalization, only 66 genes showed differentially expressed between control and MK2206 groups with >1.5-fold changes (p < 0.05, Fig. 4f). Pathway analysis revealed genes involving in ROS, and hypoxia which were ranked at the top by GO term in MK2206 treated ULM (Fig. 4g).
Molecular and cellular analysis of MK2206 induced senescence in primary ULM cells. a Immunohistochemistry analysis for pAKT in ULM spheroids (left) showed significant loss of pAKT after MK2206 treatment (right). b Primary ULM cells were treated with 5 μM MK2206 or the vehicle (DMSO) and cells from 2D and 3D were stained for SA-β-gal and the dark blue (2D) and green (3D) stains were indicative of senescent cells. c Immunohistochemistry revealed increase of p16 and p21 expression in MK2206-treated ULM spheroids and a total of 8 spheroids were scored (right). d Western blot analysis for pAKT, total AKT, P21, and β-actin. Densitometric analysis was done (n = 4). e Differentially expressed genes in MK2206- vs DMSO-treated ULM were plotted in heatmap and Dendragram where upregulated genes are red and downregulated genes are blue. f GO analysis showed four highly upregulated functional pathways in ULM treated by MK2206. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Functional analysis of the selected genes dysregulated in both replication- and MK2206-induced senescence
Although numerous distinct pathways are regulated by passaging vs AKT inhibition, both approaches resulted in senescence and thus we focused on the common genes between the two gene sets to potentially identify novel senescence associated genes in ULM. Four genes were found to be significantly upregulated in both replicative and MK2206 induced senescence (Fig. 5a). Validated of these genes were done in additional primary myometrial and ULM cells and WIPI1 and SLITKR4 were selected for further analysis. Western blot analysis demonstrated that both WIPI1 and SLITKR4 protein were significantly upregulated in ULM cells with replication senescence (P2 in Fig. 5b, Suppl Fig. 4) and ULM with stress-induced senescence (MK2206 in Fig. 5c, Suppl Fig. 4).
Analysis of the selected senescence-associated genes in ULM cells induced by replication and MK2206-induced senescence. a Venn diagrams demonstrate the number of unique and overlapping genes between the two groups (P2 vs. P0 in blue, and MK2206 vs. DMSO in yellow). Four genes shared by serial passaging and MK2206 treatment were SGIP1, SLITRK4, FAM108C1, and WIPI1. b, c Western blots of WIPI1 and SLITRK4 expression in replication senescent ULM cells (b, confirmed by upregulation of P21) and MK2206-induced senescence (c, confirmed by loss of pAKT). AKT and β-actin were used as loading control. The band density (right) was quantified (n = 3) and expressed as the means ± SD. d SA-β-gal staining was performed after overexpression of WIPI1 or SLITRK4 in 2D and 3D primary ULM cells. ULM spheroids are shown and color intensity was quantified with Image J for WIPI1 and SLITRK4. *p < 0.05, **p < 0.01, ***p < 0.001
To investigate the functional role of these two genes, WIPI1 and SLITRK4 genes were overexpressed (Suppl Figs. 5A, 5B) and senescence of ULM from 2D and 3D culture were examined by SA-β-gal staining. As shown in Fig. 5d, overexpression of WIPI1 and SLITRK4 resulted in significant cellular senescence in ULM cells detected by SA-β-gal staining intensity. In contrast to P2 or MK2206-induced senescence, in which diffuse SA-β-gal staining pattern in almost all 3D nodules (Figs. 3a, 4b), senescence in WIPI1 and SLITRK4-transfected ULM cells showed patchy and uneven distribution. This is likely due to the varied transfection efficacy of WIPI1 and SLITRK4 in different cells and gave rise to a different SA-β-gal staining pattern in 3D model. Nevertheless, overexpression of these two genes induced cellular senescence in ULM cells. We further examined p16 and p21 expression in ULM cells with and without WIPI1 and SLITRK4 overexpression. Interestingly, WIPI1 overexpression increased p21 but not p16 expression, and SLITRK4 overexpression increased p16 but p21 expression (Suppl Fig. 5). This finding suggests that these two genes are downstream AKT effectors to regulate different cell cycle inhibitors.
Senolytic analysis of anti-bcl2 treatment in replication and MK2206-induced senescence
Accumulating evidence has shown that the BCL2 inhibitors act as senolytic agents in that they can specifically target senescent cells and promote death. Given that ULM undergo both replicative and stress-induced senescence, we tested whether the BCL2 inhibitor, ABT263, could clear senescent ULM cells. ULM cells were passaged twice (P2) to reach replication senescence as illustrated in Fig. 3a. Spheroids of P2 ULM were treated with ABT263. ABT263 treatment resulted in rim of disassociated cells (Fig. 6a upper panel) and histologic evaluation revealed that increased apoptosis/cell death in spheroids treated with ABT263 as shown by vacuolated cells and pyknotic nuclei (Fig. 6a lower panel). To confirm that ABT263 effectively removed senescent cells, SA-β-gal staining was performed in P2 myometrial and ULM spheroids which were treated with DMSO control and ABT263. As the intensity of green color represented the level of cellular senescence in spheroids, both myometrial and ULM spheroids showed loss of senescent cells in ABT263 treatment (Fig. 6b). Next, we prepared P0 spheroids from another 3 ULMs and matched myometrium. Spheroids were treated with control (DMSO), ABT263, MK2206 and ABT263 + MK2206. Administration of ABT263 can effectively depleted senescent cells from nature occurring and MK2206-induced senescence (Fig. 6c). When comparing the senolytic effect by ABT263 from day 1 to day 3, further reduction of senescent cells from day 3 were observed (Fig. 6d, Suppl Fig. 6). To confirm the findings, ULM cells were prepared in 2D culture and treated with same condition as illustrated in Fig. 3, cell viability were examined by counting attached living cells (Fig. 6e) and similar results were obtained as seen in spheroid culture. Our results demonstrate that selective clearance of senescent cells by ABT263 is reproducible in 3D ULM tumor nodule and senolytic drugs may be considered to be potential agents for further analysis.
Depletion of senescent cells by ABT263, a senolytic reagent, in primary ULM spheroids. a Primary ULM cells were serially passaged (P2), prepared in 3D spheroids and treated with ABT263. Top panels showed cell death to form a rim of dissociated cells around spheroids. Bottom panels showed apoptosis cells in spheroids prepared in formalin-fixed and paraffin-embedded section. b P2 ULM spheroids were treated with DMSO and ABT263 and senescence were examined by SA-β-gal stain. Intensity of blue color represented the level of senescence. c ULM spheroids were treated with DMSO, ABT263, MK2206 and MK2206 + ABT263 and examined by SA-β-gal stain and the level of senescence were measured based on the intensity of green color (right panel, n = 3). d Senescent rate were measure in primary ULM spheroids from day 1 to day 3 based on intensity of SA-β-gal stain. e Cell viability analysis in 2D culture in primary ULM cells of control (DMSO), ABT263, MK2206 and MK2206 + ABT263. Cell counts were measured and illustrated on right panel with the means ± sem. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Discussion
The model of ex vivo three-dimensional (3D) spheroids is a new and valuable tool for ULM study [13]. It architecturally resembles ULM in vivo, characterized by maintaining the cellular and molecular features similar to primary ULM. It overcomes the limitations of 2D ULM culture, in which ULM cells quickly lose expression of smooth muscle actin and sex steroid hormone receptors. It can be an attractive method, in which establishing the spheroid tumor nodules from primary tumor cells can be mimicked to in vivo cyto-morphologic nature of given tumors in short period of time and can be used for functional assays and drug testing within a short time frame, as a comparison to ULM xenograft models [14, 15]. In particular, a 3D model can be easily accessed for the analysis of molecular and cytopathologic changes in replication and stress-induced senescence (Figs. 3, 4). In addition, ULM spheroids provide enough material for cytopathologic and molecular analysis through serial sections as illustrated in Figs. 1 and 2. For example, spheroids from one patient ULM was used to evaluate gene expression, and immunohistochemistry (Figs. 2, 4). Thus, 3D spheroid cultures for ULM is a promising tool for ex vivo ULM studies, particularly for testing therapeutic modalities.
Naturally occurring or stressed-induced cellular senescence are common and important biologic phenotypes in primary ULM. Study of cellular senescence in ULM will greatly facilitate understanding the molecular and cellular mechanisms for potential therapeutic targets. Cellular senescence is usually characterized by permanent growth arrest with a failure to re-enter the cell cycle. The cyto-morphologic features of senescent cells often include large, multinucleated, and vacuolated appearances [16, 17]. The molecular biomarkers for cellular senescence include the presence of senescence-associated β-galactosidase (SA-β-gal) at pH6.0, the increased expression of p16 and p21, the secretion of senescence-associated secretory phenotype (SASP), the appearance of senescence-associated heterochromatic foci (SAHF) and DNA damage foci (DNA-SCARS/TIF) [18]. ULM are slow growing benign smooth muscle tumors which can undergo spontaneous growth arrest or senescence given the stress conditions [5]. It has been suggested that senescence, which may be responsible for growth restrictions frequently observed in ULM, may efficiently protect them against malignant transformation [6, 19]. Moreover, several compounds, including MK2206, an allosteric AKT inhibitor, and nutlin-3, a MDM2 inhibitor, can induce senescence in uterine leiomyoma [7, 20]. Therefore, investigating the molecular pathways and identifying the novel senescence associated biomarkers specific to ULM can provide us an insight into the development of more effective therapeutics against the disease.
We previously found a varied level of cellular senescence naturally occurring in nearly 50% of ULM [5]. This is consistent with the findings in which, a proportion of ULM do not grow by a long follow-up study [19]. Therefore, characterization of the molecular change in cellular senescence will greatly facilitate our understanding the pathophysiology of ULM. Although permanent cell cycle arrest would be a desired response to prevent ULM growth and decrease the risk for malignant transformation, there is accumulating evidence that senescent cells can have deleterious effects on the tissue microenvironment, especially in terms of tumor growth [21]. The transition of a cell to a SASP is characterized by changes in protein expression and secretion and cell proliferation. Some of these secreted factors, including interleukins, chemokines, growth factors, and inflammatory mediators, are pro-mitogenic, which can affect surrounding tumor cells [21, 22]. The importance of senescence in ULM and its therapeutic potential remains to be elucidated.
In this study, we examined and evaluate the replication and stress-induced senescence in ULM with ex vivo 3D spheroid model. Replication senescence is induced by serial passaging and stress-induced senescence is induced by inhibiting AKT. These two senescence may involve different mechanisms at upstream molecular reaction and may converge on some common pathways at downstream that lead to senescence. This is demonstrated by the large number of differentially expressed genes in the cells that were serially passaged compared to the number of genes in the MK2206-treated cells (Fig. 2 and published data [23, 24]). The similar senescent phenotypes presented in both groups suggest that a common group of target genes may be required despite the different pathways. Our pathway analysis revealed that the pathways involved by the dysregulated genes are broad and 11 common pathways shared by passage and MK2206-treated ULM cells, including autoimmune and inflammatory network, cell adhesion and smooth muscle function (Fig. 2).
Global gene expression analysis by comparing two senescence in ULM, we identified two novel target genes (WIPI1 and SLITRK4) that were upregulated in both senescence pathway we focused on identifying genes that were shared by both pathways (Fig. 5). WIPI1 and SLITRK4 were consistently upregulated in ULM cells undergoing replication or stress-induced senescence. WIPI1 (WD repeat domain, phosphoinositide interacting 1, ATG18 protein homolog) has been demonstrated to be involved in starvation- and calcium-mediated autophagy [25]. It is also known to induce cellular senescence by transactivating p21 [26]. Yang et al. [27] reported that WIPI1 was upregulated in doxorubicin treated senescent K562 cells. In autophagy, WIPI1 is recruited early to form autophagosomes [28] and plays an important role in vesicle trafficking in cells [29], contributing to the secretion of SASP [30]. Hence, upregulated WIPI1 is suggestive of the initiation of autophagy in senescent primary uterine leiomyoma cells, which is consistent with our previous finding that autophagy is activated during the process of senescence [31]. Moreover, Johmura et al. [26]. demonstrated that WIPI1 was markedly induced in the senescent populations upon ionizing radiation treatment of normal human fibroblast HCA2 cells. WIPI1 was also shown to transactivate p21 [14]. SLITRK4 (SLIT and NTRK like family member 4) is a member of the SLITRK family that is predominantly expressed in neural tissues and it suppresses neurite outgrowth [32]. The function of SLITRK4 in senescence is less clear. SLITRK4 expression was significantly reduced in breast cancer tissue in comparison to normal breast tissue [33]. According to Human Protein Atlas (HPA), both WIPI1 and SLITRK4 expression are relatively high in human uterus in comparison to other organ system, suggesting their specific role in uterine function. In our study, overexpression of either WIPI1 or SLITRK4 induced senescence in ULM and myometrial (data not shown) cells demonstrating their major role in cellular senescence. Further characterization of molecular mechanism of these two gene expression and functional pathway will greatly facilitate in development of new therapeutic modality targeting at downstream and possible specific functional pathway in controlling ULM growth and senescence.
In summary, we have systematically evaluated the methods and cytohistologic features of 3D spheroid ULM model in the analysis of cellular senescence. We identified two novel senescence-associated genes in ULM, WIPI1 and SLITRK4 which also have the ability to induce senescence. Additional investigation of senescence associated gene functions induced by aging and AKT stress may provide new tools for understanding of the senescence mechanisms and future target therapies.
References
Bulun SE. Uterine fibroids. N Engl J Med. 2013;369:1344–55.
Ciavattini A, Di Giuseppe J, Stortoni P, Montik N, Giannubilo SR, Litta P, et al. Uterine fibroids: pathogenesis and interactions with endometrium and endomyometrial junction. Obstet Gynecol Int. 2013;2013:173184.
Chabbert-Buffet N, Esber N, Bouchard P. Fibroid growth and medical options for treatment. Fertil Steril. 2014;102:630–9.
Mehine M, Kaasinen E, Heinonen HR, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci USA. 2016;113:1315–20.
Laser J, Lee P, Wei JJ. Cellular senescence in usual type uterine leiomyoma. Fertil Steril. 2010;93:2020–6.
Markowski DN, Helmke BM, Belge G, et al. HMGA2 and p14Arf: major roles in cellular senescence of fibroids and therapeutic implications. Anticancer Res. 2011;31:753–61.
Xu X, Lu Z, Qiang W, et al. Inactivation of AKT induces cellular senescence in uterine leiomyoma. Endocrinology. 2014;155:1510–9.
Vidimar V, Gius D, Chakravarti D, et al. Dysfunctional MnSOD leads to redox dysregulation and activation of prosurvival AKT signaling in uterine leiomyomas. Sci Adv. 2016;2:e1601132.
McMillen BD, Aponte MM, Liu Z, et al. Expression analysis of MIR182 and its associated target genes in advanced ovarian carcinoma. Mod Pathol. 2012;25:1644–53.
Wagner W, Bork S, Horn P, et al. Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS ONE. 2009;4:e5846.
Johmura Y, Nakanishi M. Multiple facets of p53 in senescence induction and maintenance. Cancer Sci. 2016;107:1550–5.
Post WS, Goldschmidt-Clermont PJ, Wilhide CC, et al. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999;43:985–91.
Vidimar V, Chakravarti D, Bulun SE, et al. The AKT/BCL-2 axis mediates survival of uterine leiomyoma in a novel 3D spheroid model. Endocrinology 2018.
Ishikawa H, Ishi K, Serna VA, et al. Progesterone is essential for maintenance and growth of uterine leiomyoma. Endocrinology. 2010;151:2433–42.
Qiang W, Liu Z, Serna VA, et al. Down-regulation of miR-29b is essential for pathogenesis of uterine leiomyoma. Endocrinology. 2014;155:663–9.
Althubiti M, Lezina L, Carrera S, et al. Characterization of novel markers of senescence and their prognostic potential in cancer. Cell Death Dis. 2014;5:e1528.
Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40.
Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–56.
Peddada SD, Laughlin SK, Miner K, et al. Growth of uterine leiomyomata among premenopausal black and white women. Proc Natl Acad Sci USA. 2008;105:19887–92.
Markowski DN, Helmke BM, Radtke A, et al. Fibroid explants reveal a higher sensitivity against MDM2-inhibitor nutlin-3 than matching myometrium. BMC Women’s Health. 2012;12:2.
Coppe JP, Desprez PY, Krtolica A, et al. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118.
Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94.
Kennedy AL, Morton JP, Manoharan I, et al. Activation of the PIK3CA/AKT pathway suppresses senescence induced by an activated RAS oncogene to promote tumorigenesis. Mol Cell. 2011;42:36–49.
Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68.
Grotemeier A, Alers S, Pfisterer SG, et al. AMPK-independent induction of autophagy by cytosolic Ca2+ increase. Cell Signal. 2010;22:914–25.
Johmura Y, Sun J, Kitagawa K, et al. SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat Commun. 2016;7:10574.
Yang MY, Lin PM, Liu YC, et al. Induction of cellular senescence by doxorubicin is associated with upregulated miR-375 and induction of autophagy in K562 cells. PLoS ONE. 2012;7:e37205.
Tsuyuki S, Takabayashi M, Kawazu M, et al. Detection of WIPI1 mRNA as an indicator of autophagosome formation. Autophagy. 2014;10:497–513.
Ho H, Kapadia R, Al-Tahan S, et al. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J Biol Chem. 2011;286:12509–23.
Urbanelli L, Buratta S, Sagini K, et al. Extracellular vesicles as new players in cellular senescence. Int J Mol Sci. 2016;17.
Galluzzi L, Bravo-San Pedro JM, Kroemer G. Autophagy mediates tumor suppression via cellular senescence. Trends Cell Biol. 2016;26:1–3.
Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci. 2003;24:117–29.
Haakensen VD, Bjoro T, Luders T, et al. Serum estradiol levels associated with specific gene expression patterns in normal breast tissue and in breast carcinomas. BMC Cancer. 2011;11:332.
Acknowledgements
This study was supported by NIH P01HD57877. This work was also partially supported by the Northwestern University Pathology Core Facility and NUSeq core for the gene profiling studies from a Cancer Center Support Grant (NCI CA060553).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Xie, J., Xu, X., Yin, P. et al. Application of ex-vivo spheroid model system for the analysis of senescence and senolytic phenotypes in uterine leiomyoma. Lab Invest 98, 1575–1587 (2018). https://doi.org/10.1038/s41374-018-0117-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0117-5