Abstract
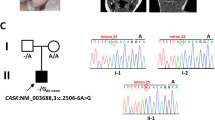
Ubiquitin-specific protease 8 (USP8) is a deubiquitinating enzyme involved in deubiquitinating the enhanced epidermal growth factor receptor for escape from degradation. Somatic variants at a hotspot in USP8 are a cause of Cushing’s disease, and a de novo germline USP8 variant at this hotspot has been described only once previously, in a girl with Cushing’s disease and developmental delay. In this study, we investigated an exome-negative patient with severe developmental delay, dysmorphic features, and multiorgan dysfunction by long-read sequencing, and identified a 22-kb de novo germline deletion within USP8 (chr15:50469966-50491995 [GRCh38]). The deletion involved the variant hotspot, one rhodanese domain, and two SH3 binding motifs, and was presumed to be generated through nonallelic homologous recombination through Alu elements. Thus, the patient may have perturbation of the endosomal sorting system and mitochondrial autophagy through the USP8 defect. This is the second reported case of a germline variant in USP8.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Mevissen TET, Komander D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem. 2017;86:159–92.
Schauer NJ, Magin RS, Liu X, Doherty LM, Buhrlage SJ. Advances in Discovering Deubiquitinating Enzyme (DUB) Inhibitors. J Med Chem. 2020;63:2731–50.
Dufner A, Knobeloch KP. Ubiquitin-specific protease 8 (USP8/UBPy): a prototypic multidomain deubiquitinating enzyme with pleiotropic functions. Biochem Soc Trans. 2019;47:1867–79.
Durcan TM, Tang MY, Perusse JR, Dashti EA, Aguileta MA, McLelland GL, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014;33:2473–91.
Pearson G, Soleimanpour SA. A ubiquitin-dependent mitophagy complex maintains mitochondrial function and insulin secretion in beta cells. Autophagy. 2018;14:1160–61.
Pearson G, Soleimanpour SA. Visualization of Endogenous Mitophagy Complexes In Situ in Human Pancreatic Beta Cells Utilizing Proximity Ligation Assay. J Vis Exp. 2019;147:10.3791/59398.
Kakihara K, Asamizu K, Moritsugu K, Kubo M, Kitaguchi T, Endo A, et al. Molecular basis of ubiquitin-specific protease 8 autoinhibition by the WW-like domain. Commun Biol. 2021;4:1272.
Schoneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol. 2017;18:5–17.
McCullough J, Frost A, Sundquist WI. Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes. Annu Rev Cell Dev Biol. 2018;34:85–109.
Reincke M, Sbiera S, Hayakawa A, Theodoropoulou M, Osswald A, Beuschlein F, et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet. 2015;47:31–8.
Theodoropoulou M, Reincke M. Genetics of Cushing’s disease: from the lab to clinical practice. Pituitary. 2022;25:689–92.
Ma ZY, Song ZJ, Chen JH, Wang YF, Li SQ, Zhou LF, et al. Recurrent gain-of-function USP8 mutations in Cushing’s disease. Cell Res. 2015;25:306–17.
Cohen M, Persky R, Stegemann R, Hernandez-Ramirez LC, Zeltser D, Lodish MB, et al. Germline USP8 Mutation Associated With Pediatric Cushing Disease and Other Clinical Features: A New Syndrome. J Clin Endocrinol Metab. 2019;104:4676–82.
Sakamoto M, Shiiki T, Matsui S, Okamoto N, Koshimizu E, Tsuchida N, et al. A novel homozygous CHMP1A variant arising from segmental uniparental disomy causes pontocerebellar hypoplasia type 8. J Hum Genet. 2023;68:247–53.
Fromer M, Purcell SM. Using XHMM Software to Detect Copy Number Variation in Whole-Exome Sequencing Data. Curr Protoc Hum Genet. 2014;81::7 23 1–7 23 21.
Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12:184.
Mori T, Sakamoto M, Tayama T, Goji A, Toda Y, Fujita A, et al. A case of epilepsy with myoclonic atonic seizures caused by SLC6A1 gene mutation due to balanced chromosomal translocation. Brain Dev. 2023;45:395–400.
Mitsuhashi S, Ohori S, Katoh K, Frith MC, Matsumoto N. A pipeline for complete characterization of complex germline rearrangements from long DNA reads. Genome Med. 2020;12:67.
Pearson G, Chai B, Vozheiko T, Liu X, Kandarpa M, Piper RC, et al. Clec16a, Nrdp1, and USP8 Form a Ubiquitin-Dependent Tripartite Complex That Regulates beta-Cell Mitophagy. Diabetes. 2018;67:265–77.
Row PE, Liu H, Hayes S, Welchman R, Charalabous P, Hofmann K, et al. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem. 2007;282:30929–37.
Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–74.
Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803.
Mauri S, Bernardo G, Martinez A, Favaro M, Trevisan M, Cobraiville G, et al. USP8 Down-Regulation Promotes Parkin-Independent Mitophagy in the Drosophila Brain and in Human Neurons. Cells 2023;12:1143.
Wiszniewski W, Hunter JV, Hanchard NA, Willer JR, Shaw C, Tian Q, et al. TM4SF20 ancestral deletion and susceptibility to a pediatric disorder of early language delay and cerebral white matter hyperintensities. Am J Hum Genet. 2013;93:197–210.
Hjeij R, Lindstrand A, Francis R, Zariwala MA, Liu X, Li Y, et al. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am J Hum Genet. 2013;93:357–67.
Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, Baggett BC, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am J Hum Genet. 2014;95:143–61.
de Smith AJ, Walters RG, Coin LJ, Steinfeld I, Yakhini Z, Sladek R, et al. Small deletion variants have stable breakpoints commonly associated with alu elements. PLoS One. 2008;3:e3104.
Uchiyama Y, Yamaguchi D, Iwama K, Miyatake S, Hamanaka K, Tsuchida N, et al. Efficient detection of copy-number variations using exome data: Batch- and sex-based analyses. Hum Mutat. 2021;42:50–65.
Acknowledgements
This work was supported by AMED under grant numbers JP23ek0109674, JP23ek0109549, JP23ek0109617, JP23ek0109648 (NM); JSPS KAKENHI under grant numbers JP23H02829 (SM), JP23H02877 (TM), JP22K15901 (AF), JP21K07869 (EK), JP23K07229 (YU), and JP23K15353 (NT); and the Takeda Science Foundation (TM and NaM). We thank Catherine Perfect, MA (Cantab), from Edanz (https://jp.edanz.com/ac), for editing a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: MS, NM; Data Curation: MS, KI; Investigation: MS, KK, KT, FI, YW, NO; Visualization and Writing-original draft: MS; Writing-review and editing: NT, YU, EK, AF, KM, SM, TM, NM.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sakamoto, M., Kurosawa, K., Tanoue, K. et al. A heterozygous germline deletion within USP8 causes severe neurodevelopmental delay with multiorgan abnormalities. J Hum Genet 69, 85–90 (2024). https://doi.org/10.1038/s10038-023-01209-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01209-2