Abstract
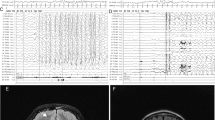
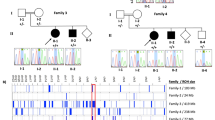
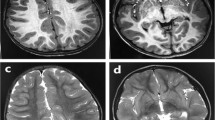
The ubiquitin–proteasome system is the principal system for protein degradation mediated by ubiquitination and is involved in various cellular processes. Cullin-RING ligases (CRL) are one class of E3 ubiquitin ligases that mediate polyubiquitination of specific target proteins, leading to decomposition of the substrate. Cullin 3 (CUL3) is a member of the Cullin family proteins, which act as scaffolds of CRL. Here we describe three cases of global developmental delays, with or without epilepsy, who had de novo CUL3 variants. One missense variant c.854T>C, p.(Val285Ala) and two frameshift variants c.137delG, p.(Arg46Leufs*32) and c.1239del, p.(Asp413Glufs*42) were identified by whole-exome sequencing. The Val285 residue located in the Cullin N-terminal domain and p.Val285Ala CUL3 mutant showed significantly weaker interactions to the BTB domain proteins than wild-type CUL3. Our findings suggest that de novo CUL3 variants may cause structural instability of the CRL complex and impairment of the ubiquitin–proteasome system, leading to diverse neuropsychiatric disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Hershko A, Ciechanover A. Mechanisms of intracellular protein breakdown. Annu Rev Biochem. 1982;51:335–64.
Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–51.
Segref A, Hoppe T. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep. 2009;10:44–50.
Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428.
Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220.
Huber C, Dias-Santagata D, Glaser A, O’Sullivan J, Brauner R, Wu K, et al. Identification of mutations in CUL7 in 3-M syndrome. Nat Genet. 2005;37:1119–24.
Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102.
Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–6.
Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–3.
RK CY, Merico D, Bookman M, J LH, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602–11.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–5.
Wang, TY, Guo, H, Xiong, B, Stessman, HAF, Wu, HD, Coe, BP et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. 2016;7:13316.
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50.
Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–84.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8.
Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6.
Agbor LN, Ibeawuchi SC, Hu C, Wu J, Davis DR, Keen HL, et al. Cullin-3 mutation causes arterial stiffness and hypertension through a vascular smooth muscle mechanism. JCI Insight. 2016;1:e91015.
Nagasaki M, Yasuda J, Katsuoka F, Nariai N, Kojima K, Kawai Y, et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–53.
Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Krek W. BTB proteins as henchmen of Cul3-based ubiquitin ligases. Nat Cell Biol. 2003;5:950–1.
Cullinan SB, Gordan JD, Jin JO, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–86.
Jonsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549:519–22.
Casas-Alba D, Vila Cots J, Monfort Carretero L, Martorell Sampol L, Zennaro MC, Jeunemaitre X, et al. Pseudohypoaldosteronism types I and II: little more than a name in common. J Pediatr Endocrinol Metab. 2017;30:597–601.
Glover M, Ware JS, Henry A, Wolley M, Walsh R, Wain LV, et al. Detection of mutations in KLHL3 and CUL3 in families with FHHt (familial hyperkalaemic hypertension or Gordon’s syndrome). Clin Sci (Lond). 2014;126:721–6.
Mori T, Hosomichi K, Chiga M, Mandai S, Nakaoka H, Sohara E, et al. Comprehensive genetic testing approach for major inherited kidney diseases, using next-generation sequencing with a custom panel. Clin Exp Nephrol. 2017;21:63–75.
Osawa M, Ogura Y, Isobe K, Uchida S, Nonoyama S, Kawaguchi H. CUL3 gene analysis enables early intervention for pediatric pseudohypoaldosteronism type II in infancy. Pediatr Nephrol. 2013;28:1881–4.
Shao L, Cui L, Lu J, Lang Y, Bottillo I, Zhao X. A novel mutation in exon 9 of Cullin 3 gene contributes to aberrant splicing in pseudohypoaldosteronism type II. FEBS Open Bio. 2018;8:461–9.
Ellison DH. Pseudohypoaldosteronism Type II. 2011 Nov 10 [Updated 2017 Feb 16]. In Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K et al. editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020.
Healy JK. Pseudohypoaldosteronism type II: history, arguments, answers, and still some questions. Hypertension. 2014;63:648–54.
Anderica-Romero AC, Gonzalez-Herrera IG, Santamaria A, Pedraza-Chaverri J. Cullin 3 as a novel target in diverse pathologies. Redox Biol. 2013;1:366–72.
Schumacher FR, Siew K, Zhang J, Johnson C, Wood N, Cleary SE, et al. Characterisation of the Cullin-3 mutation that causes a severe form of familial hypertension and hyperkalaemia. EMBO Mol Med. 2015;7:1285–306.
Ferdaus, MZ, Miller, LN, Agbor, LN, Saritas, T, Singer, JD, Sigmund, CD et al. Mutant Cullin 3 causes familial hyperkalemic hypertension via dominant effects. JCI Insight. 2017;2. pii: 96700. https://doi.org/10.1172/jci.insight.96700.
Ibeawuchi SR, Agbor LN, Quelle FW, Sigmund CD. Hypertension-causing mutations in Cullin3 protein impair RhoA protein ubiquitination and augment the association with substrate adaptors. J Biol Chem. 2015;290:19208–17.
Gordon RD. Syndrome of hypertension and hyperkalemia with normal glomerular filtration rate. Hypertension. 1986;8:93–102.
Guo H, Wang T, Wu H, Long M, Coe BP, Li H, et al. Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Mol Autism. 2018;9:64.
Codina-Sola M, Rodriguez-Santiago B, Homs A, Santoyo J, Rigau M, Aznar-Lain G, et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol Autism. 2015;6:21.
McCormick JA, Yang CL, Zhang C, Davidge B, Blankenstein KI, Terker AS, et al. Hyperkalemic hypertension-associated cullin 3 promotes WNK signaling by degrading KLHL3. J Clin Invest. 2014;124:4723–36.
Agbor, LN, Nair, AR, Wu, J, Lu, KT, Davis, DR, Keen, HL et al. Conditional deletion of smooth muscle Cullin-3 causes severe progressive hypertension. JCI Insight. 2019;5. pii: 129793. https://doi.org/10.1172/jci.insight.129793.
Rapanelli M, Tan T, Wang W, Wang X, Zhong P, Wang ZJ, et al. Behavioral, circuitry and molecular aberrations by region-specific deficiency of the high-risk autism gene CUL3. J Neurochem. 2019;150:187–187.
Dong Z, Chen W, Chen C, Wang H, Cui W, Tan Z, et al. CUL3 deficiency causes social deficits and anxiety-like behaviors by impairing excitation-inhibition balance through the promotion of cap-dependent translation. Neuron. 2020;105:475–90 e476.
Acknowledgements
We express our gratitude to all patients and their families for participating in this study. We would also like to thank Ms. M. Sato, N. Watanabe, M Tsujimura, K Shibasaki and A. Kitamoto for their technical assistance. This work was supported by AMED under the grant numbers JP19ek0109280, JP19dm0107090, JP19ek0109301, JP19ek0109348, JP18kk020501 (to NM); JSPS KAKENHI under the grant numbers JP16H05160 (to HS), JP16K09975 (to MK), JP17H01539 (to NM) and JP19K07977 (to SM); grants from the Ministry of Health, Labor, and Welfare (to NM and MK); and the Takeda Science Foundation (to MN, HS and NM); HUSM Grant-in-Aid from Hamamatsu University School of Medicine.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
URLs. 3.5KJPN (https://ijgvd.megabank.tohoku.ac.jp/). gnomAD (http://exac.broadinstitute.org). SIFT (http://provean.jcvi.org/index.php). Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/). CADD (http://gnomad.broadinstitute.org/). MutationTaster (http://www.mutationtaster.org/). GERP (http://mendel.stanford.edu/SidowLab/downloads/GERP/index.html). PhastCons (http://compgen.cshl.edu/phast/)
Supplementary information
Rights and permissions
About this article
Cite this article
Nakashima, M., Kato, M., Matsukura, M. et al. De novo variants in CUL3 are associated with global developmental delays with or without infantile spasms. J Hum Genet 65, 727–734 (2020). https://doi.org/10.1038/s10038-020-0758-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0758-2
This article is cited by
-
Neuronal knockdown of Cullin3 as a Drosophila model of autism spectrum disorder
Scientific Reports (2024)
-
A ubiquitin-based effector-to-inhibitor switch coordinates early brain, craniofacial, and skin development
Nature Communications (2023)
-
LZTR1 deficiency exerts high metastatic potential by enhancing sensitivity to EMT induction and controlling KLHL12-mediated collagen secretion
Cell Death & Disease (2023)
-
A novel missense variant in CUL3 shows altered binding ability to BTB-adaptor proteins leading to diverse phenotypes of CUL3-related disorders
Journal of Human Genetics (2021)
-
Cul3 regulates cytoskeleton protein homeostasis and cell migration during a critical window of brain development
Nature Communications (2021)