Abstract
Intrauterine growth retardation (IUGR) has been linked to metabolic syndrome including insulin resistance, and overexpression of suppressors of cytokine signaling (SOCSs) proteins is thought to be associated with increased whole-body insulin sensitivity. The insulin-resistant IUGR rat model was established by maternal food restriction (about 30% of the normal rats). The weight, length, and homeostasis model assessment of insulin resistance (HOMA-IR) of IUGR-born rats was higher than the control group. Insulin receptor substrate (IRS)-1 expression decreased, whereas SOCS-1 and SOCS-3 increased in the skeletal muscle of IUGR rats compared with the control group. The recombination plasmids PGPU6/GFP/Neo-SOCS-1small hairpin RNA (shRNA) and PGPU6/GFP/Neo-SOCS-3shRNA were transfected into skeletal muscle cells, and the shRNAs efficiently inhibited the expression of SOCS-1 and SOCS-3. Insulin-stimulated glucose transporter-4 (GLUT4) translocation was also dramatically increased. In conclusion, these data provide additional information on the mechanism of insulin resistance associated with IUGR. Down-regulation of SOCS-1 and SOCS-3 ameliorates the capacity of glucose transport and provides a potential gene therapy approach to managing metabolic syndrome.
Similar content being viewed by others
Main
IUGR, a disease complex that is increasing at epidemic rates worldwide, has been linked to metabolic syndrome, including insulin resistance, obesity, diabetes, dyslipidemia, and nonalcoholic fatty liver (1–3). Insulin resistance is an essential aspect of metabolic syndrome, and numerous cytokines reportedly contribute to the development of these disorders (4); however, the molecular mechanisms leading to the development of metabolic syndrome in intrauterine growth retardation (IUGR) -born adults is still poorly understood.
Suppressors of cytokine signaling (SOCSs) are thought to participate in negative feedback loops in cytokine signaling via various mechanisms. A promoter polymorphism of the SOCS-3 gene is thought to be associated with increased whole-body insulin sensitivity. One study has shown that overexpression of either SOCS-1 or SOCS-3 attenuates insulin-induced glycogen synthesis in L6 myotubes and activation of glucose uptake in 3T3L1 adipocytes (5). Recent studies showed that SOCS-1 knockout mice had lower glucose levels than WTs and that cells derived from the SOCS-1 knockout mice apparently exhibited enhanced insulin signaling. However, it is important to note that determining insulin sensitivity in vivo using SOCS-1 knockout mice is difficult because they die within 3 wk of birth (6).
In this study, we investigated basal SOCS-1, SOCS-3, glucose transporter-4 (GLUT4), and insulin receptor substrate (IRS)-1 content in insulin resistance-associated IUGR rats. And on the basis of comparing various features between IUGR and normal models, we hypothesized that down-regulation of SOCS-1 and SOCS-3 through small hairpin RNAs (shRNAs) may promote insulin-stimulated GLUT4 translocation in IUGR skeletal muscle cells.
MATERIALS AND METHODS
Animal model.
Forty adult female Sprague Dawley rats weighing 200 to 250 g and 10 adult male rats were fed. Females were caged overnight with fertile males, and the presence of sperm in a vaginal smear was designated as day 0 of pregnancy. Pregnant rats were randomly divided into the control and experimental groups. Rats in the experimental group were fed 7.5 g feedstuff per day (about 30% of the normal rats), whereas the control groups were fed a standard rodent chow. The birth weight and body length were measured at birth. Rats weighing >5.5 g were included in the control group, whereas rats weighing <5.1 g were included in the experimental group. A glucose tolerance test and insulin release test were performed on all rats at 12 wk of age. All experimental procedures were approved by the Center of Experimental Animals, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
Skeletal muscle cell culture.
Skeletal muscle cells were cultured from the quadriceps femoris of rats using standard techniques. Quadriceps femoris samples were sliced into 1 mm3 fragments, and the tissues were incubated for 4 h at 37°C in a 5% humidified CO2-controlled atmosphere. Dulbecco's modified Eagle's medium (DMEM; GIBCO), 10% heat-inactivated fetal bovine serum (GIBCO), 100 U/mL penicillin, and 100 μg/mL streptomycin were added. When cells reached 70–80% confluence, cultures were digested with 2% trypsin (GIBCO) and seeded onto six-well plates. Immunocytochemical staining was performed with antisarcomeric actin (Boster, China) in 1:100 dilution.
shRNA preparation and plasmid construction.
Rat SOCS-1, SOCS-3 gene sequences were obtained from GenBank (GI: 22024395 and 16758335). Three small interfering RNA (siRNA) sequences were designed, and one random sequence was prepared as a control. The shRNA sense oligonucleotides contained a 4-nucleotide overhang to create a BbsI restriction site (CACC) plus G, followed by a 21-nucleotide sense siRNA sequence, a 9-nucleotide loop (TTCAAGAGA), a 21-nucleotide reverse complementary antisense siRNA sequence, and a polymerase III terminator (TTTTTA). The complementary antisense oligonucleotides contained a four-nucleotide overhang at the 5′ terminus to create a BamHI restriction site. These oligonucleotides were annealed and ligated into the pSUPER vector (OligoEngine, Seattle, WA) via the BamHI and BbsI sites. The scrambled shRNA sequence (described previously) was used as a control. All plasmid vectors were purified using cesium chloride gradient centrifugation. The sequences of the shRNAs are provided in Table 1.
DNA sequences containing a small hairpin structure were synthesized, annealed, and inserted into the digested PGPU6/GFP/Neo vector with BbsI and BamHI, and the recombinant plasmid was transfected into the Escherichia coli strain DH5α (Fig. 1). Successful construction of PGPU6/GFP/Neo-SOCS-1shRNA and PGPU6/GFP/Neo-SOCS-3shRNA was confirmed by DNA sequencing.
Skeletal muscle cell cultures were transfected with plasmid vector, and fluorescence photographs were obtained. Positively transfected cells were counted, and a transfection efficiency >80% was confirmed.
Transfection and positive clone screening.
To obtain the highest transfection efficiency with low nonspecific effects, transfection conditions were optimized. Briefly, 5 μL Lipofectamine 2000 was gently mixed with 250 μL Opti-MEM I, incubated for 5 min at room temperature, and then diluted with 7.5 μL shRNA diluted in 250 μL Opti-MEM I. These oligodeoxynucleotide solutions were gently mixed and incubated for 5 min at room temperature and then subsequently mixed with diluted Lipofectamine and incubated for 20 min at room temperature. Fifty microliters of this solution was added to each well of a 96-well plate containing skeletal muscle cell cultures. Plates were gently mixed by rocking the plate back and forth and then incubated at 37°C in a CO2 incubator for 48 h. Transfection efficiency could be determined by the ratio of oligonucleotides labeled with FITC to the total number of cells.
Real-time PCR analysis.
Total RNA was isolated from cultured cells with Trizol reagent (Invitrogen Technologies) according to the manufacturer's protocol. RNA quality and quantity were evaluated by UV spectrophotometry at 260 and 280 nm (the A260 to A280 ratio of pure RNA was ∼2). Then RNA was reverse transcribed to cDNA using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer's instructions. The cDNA was either analyzed immediately or stored at −20°C until the time of analysis. Quantitative PCR amplification was performed by ABI 7500HT fast real-time PCR System (ABI Company), and the SYBR Green I fluorescent dye method was used to quantify cDNA. The sequence-specific primers were as follows: IRS-1 upper primer 5′-CCTGGAGTATTATGAGAACGAG-3′, lower primer 5′-GCCGCAATGGCAAAGTGT-3′; SOCS-1 upper primer 5′-CCGCTCCCACTCTGATTACCG-3′, lower primer 5′-CGAAGCCATCTTCACGCTGA-3′; SOCS-3 upper primer 5′-CTGAGCGTCGAGACCCAGTC-3′, lower primer 5′-AAGGTTCCGTCGGTGGTAAAG-3′. β-Actin (5′-ACGTTGACATCCGTAAAGAC-3′ and 5′- GAAGGTGGACAGTGAGGC-3′) was used as the internal control. All primers were synthesized by Shanghai Bio-engineering Co., Ltd., China. The final volume of the PCR was 25 μL and includes SYBR Premix Ex Taq 12.5 μL, primer (10 μM) 1 μL each, and template 1 μL. Optimization conditions were as follows: 5 min at 95°C, 40 cycles of 20 s at 94°C, annealing for 20 s at 55°C, extension for 20 s at 72°C, and a final extension for 5 min at 72°C using the Eppendorf PCR System (Eppendorf, Germany). A stable and reliable standard curve was established using synthesized oligonucleotides resembling cDNA fragments in 5-fold decrements as template. β-Actin from the same sample was used as an internal control, and the relative copy numbers of the target gene's mRNA were calculated to determine gene expression. The specificity of each reaction was controlled by melting curve analysis. Quantification was achieved using the comparative CT method, and the target transcription was normalized to an endogenous reference (β-actin). Real-time PCR was conducted in triplicate in three independent experiments.
Western blot analysis.
Proteins were extracted from cells using a Total Protein Extraction Kit (ProMab) or membrane protein extraction kit (BestBio, China). Protein concentrations were determined by the BCA assay according to the manufacturer's instructions (Pierce Co). Equal quantities of proteins (75 mg per lane) were separated by electrophoresis on a 12% SDS polyacrylamide gel and transferred onto nitrocellulose membranes. Immunoblots were sequentially incubated in 5% skimmed milk blocking solution at room temperature for 2 h. The membranes were incubated with the primary antibodies (dilutions: goat polyclonal anti-SOCS-1 and anti-SOCS-3, 1:200; polyclonal anti-IRS-1, 1:1000; rabbit polyclonal anti-GLUT4, 1:1000; rabbit polyclonal anti-Akt and anti-p-Akt-Ser473, 1:500; and rabbit polyclonal anti-GAPDH 1:1000) overnight at 4°C. After washing three times with 1% PBS-Tween solution at room temperature for 1 h, the membranes were incubated with horseradish peroxidase-conjugated IgG (1:4000; Oncogene) at room temperature for 2 h and visualized using an ECL kit. The relative amount of immunoreactive SOCS in each sample was quantified by densitometry (AMBIS Radioanalytic and Visual Imaging system, Ambis, Inc.). GAPDH was used as a protein loading control. All tests were performed in duplicate.
Immunofluorescence and confocal laser scanning microscopy.
Cultured skeletal muscle cells were transfected with the oligonucleotides at a final concentration of 500 nM in serum-free DMEM. After being cultured in DMEM plus 10% fetal bovine serum for 48 h, the cells were serum starved for 4 h and then stimulated with 0, 10−11, 10−9, and 10−7 mol/L insulin for 24 h at 37°C. Cells were washed three times with PBS for 15 min, fixed in ice-cold paraformaldehyde for 30 min, and washed three times with PBS for 15 min. Cells were incubated with primary antibody at a dilution of 1:100 for GLUT4 (Abcam, United Kingdom) overnight at 4°C and then incubated for 2 h at room temperature with FITC-conjugated goat anti-rabbit-IgG (1:1000; Oncogene). After a further washing step, 4′, 6-diamidino-2-phenylindole (DAPI) nuclear staining was used to quantify cell numbers. After 12 h, confocal microscopy was performed with a microscope (BA400, EF-UPR, Changchun, China) equipped with an argon/krypton laser. Two-channel image recording at 480 and 560 nm laser excitation was used. Optical filters were chosen for the FITC and Tetramethylrhodamine-5-(and 6)-isothiocyanate (TRLTC) range. All optical sections were recorded with the same laser and detector settings (Motic, fluo1.0). Further image processing was achieved with an image acquisition system (cooled color CCD, Nikon, Japan), and confocal stacks of green and blue fluorescence were visualized in section view mode.
Statistical analysis.
All data were expressed as mean ± SD (x̄ ± s). Differences between the two groups were compared by t test, and comparison of groups was performed via one-way ANOVA and the Student-Newman-Keuls test. Statistical analysis was performed using SPSS 7.0 software package, and a p value <0.05 was considered statistically significant.
RESULTS
Animal model.
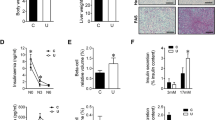
At 12 wk of age, the length of the IUGR rats was not significantly different from the control rats, but the body weights, BMI of the IUGR were significantly higher than the control group (p < 0.05, Table 2). The glucose tolerance test showed that glucose and insulin levels measured at 30, 60, 120, and 180 min were significantly higher in the IUGR group than the control group (p < 0.05, Tables 3). There was significant difference between IUGR and control group on homeostasis model assessment of insulin resistance (HOMA-IR; p < 0.05). SOCS-1 mRNA (4.3696 ± 0.8178, p < 0.05), and SOCS-3 mRNA (4.6451 ± 0.0810, p < 0.05) measured in the quadriceps femoris was significantly increased compared with the control group (1.3866 ± 0.2204, 1.0198 ± 0.0280, respectively, Table 4). Similarly, the protein levels of both SOCS-1 and SOCS-3 in the quadriceps femoris muscle were higher in the IUGR group than the control group (p < 0.05, Fig. 2) In contrast, a decline in skeletal muscle basal GLUT4 and IRS-1 protein concentration was noted in IUGR groups (p < 0.05, Fig. 2). However, there was no significant difference in total Akt between IUGR and control groups.
Skeletal muscle SOCS-1, SOCS-3, IRS-1 (A), and GLUT4, total Akt (B) concentrations from 12-wk-old IUGR and control rats under basal conditions. (A) Left: representative Western blots demonstrating IRS-1, SOCS-1, SOCS-3, and GAPDH. Right: corresponding densitometric analyses of the protein bands of interest/GAPDH shown as relative A level and presented as means (SD); (B) left: representative Western blots demonstrating GLUT4, total Akt, and GAPDH. Right: corresponding densitometric analyses of the protein bands of interest/GAPDH shown as relative A level and presented as means (SD); n = 6 per group. Open bar, control group; black bar, IUGR group. *p < 0.05 vs control group.
Down-regulation of SOCS expression of skeletal muscle cells.
The SOCS-1 and SOCS-3 mRNAs in four shRNA-treated group, one blank group, and one negative group were analyzed by real-time PCR (Figs. 3 and 4), and a significant difference between these groups was noted, which is consistent with protein levels of SOCS-1 and SOCS-3 measured by Western blot (Fig. 5). Based on these data, the most effective shRNAs were SOCS1 shRNA-b and SOCS3 shRNA-a, which were used in the subsequent experiment.
Relative expression levels of SOCS-1 (A) and SOCS-3 (B) from 12-wk-old IUGR skeletal muscle cells transfected with SOCS-1-shRNA-b and SOCS-3-shRNA-a detected by real-time PCR. n = 6 per group per treatment. Open bar, blank group; gray bar (a, b, c, and d), four shRNA-treated groups; black bar, negative group. *p < 0.05 vs blank group; §p < 0.05 vs negative group.
Relative expression levels of SOCS-1 (A) and SOCS-3 (B) from 12-wk-old IUGR skeletal muscle cells transfected with SOCS-1-shRNA-b and SOCS-3-shRNA-a detected by Western blot. Left: representative Western blots demonstrating SOCS-1 (A) and SOCS-3 (B). Right: corresponding densitometric analyses of the protein bands of interest/GAPDH shown as relative A level and presented as means (SD); n = 6 per group per treatment. Open bar, blank group; gray bar (a, b, c, and d), four shRNA-treated groups; black bar, negative group. *p < 0.05 vs blank group; §p < 0.05 vs negative group.
Representative immunofluorescence staining of GLUT4 from 12-wk-old IUGR skeletal muscle cells transfected with SOCS-1-shRNA-b and SOCS-3-shRNA-a under basal and insulin-stimulated conditions. (A1–A4) IUGR skeletal muscle cells treated with insulin (0, 10−11, 10−9, and 10−7 mol/L). (B1–B4) IUGR skeletal muscle cells transfected with SOCS-1-shRNA-b and treated with insulin (0, 10−11, 10−9, and 10−7 mol/L). (C1–C4) IUGR skeletal muscle cells transfected with SOCS-3-shRNA-a and treated with insulin (0, 10−11, 10−9, and 10−7 mol/L). Magnification was ×200, scale bars in all figures indicating 25 μm.
Promoting effect of SOCS-shRNA on insulin-stimulated glucose transport.
Fig. 5 shows representative pictures of immunofluorescence staining of GLUT4 in skeletal muscle cell membranes; insulin action leading to the plasma membrane OD of GLUT4 was detected by confocal microscopy. Glucose transfer capabilities of three groups were stimulated by different concentrations of insulin. The OD of GLUT4 in the three groups was ascending along with the increased concentration of insulin.
Figure 6 demonstrates that the GLUT4 content in plasma membranes, and the phosphorylation status of Akt under the basal state and in response to insulin stimulation (10−7mol/L) in skeletal muscle transfected with SOCS-1-shRNA-b and SOCS-3-shRNA-a. GLUT4 in plasma membrane and phospho-Akt increased in all groups after insulin stimulation compared with the basal states (p < 0.05). The content of GLUT4 in plasma membranes from IUGR skeletal muscle cells transfected with SOCS-1-shRNA-b and SOCS-3-shRNA-a had no significant difference in the base state, however, both higher after stimulated with 10−7mol/L insulin compared with control group (p < 0.05). Both transfected groups show higher phosphorylation of Akt in response to insulin stimulation compared with IUGR control group, although not significantly higher under basal conditions.
GLUT4 in plasma membranes (A) and phospho-Akt (B) from 12-wk-old IUGR skeletal muscle cells transfected with SOCS-1-shRNA-b and SOCS-3-shRNA-a under basal (−) and insulin-stimulated conditions (+). Left: representative Western blots demonstrating GLUT4 in plasma membranes or phospho-Akt. Right: corresponding densitometric analyses of the protein bands of interest/GAPDH shown as relative A level and presented as means (SD); n = 6 per group per treatment. Open bar, IUGR under basal conditions; black bar, IUGR insulin-stimulated; and IUGR skeletal muscle cells treated with SOCS-1-shRNA-b or SOCS-3-shRNA-a under basal (dark gray) and insulin-stimulated (light gray) conditions. *p < 0.05 insulin stimulated vs basal conditions of the same group, §p < 0.05 vs IUGR control group insulin-stimulated.
DISCUSSION
IUGR occurs as a consequence of poor maternal nutrition, placental insufficiency, diminished fetal oxygenation, exposure to teratogens, and other causes. IUGR animal models can be created by using such methods because of maternal food restriction and dexamethasone exposure. Prenatal food restriction was a sensitive model aimed at investigating the link between LBW and development of insulin resistance later in life (7). In this study, limited maternal nutrition in Sprague Dawley rats was used to establish the animal model.
Both in animals and humans, intrauterine deprivation of nutrition programs the fetus to develop insulin resistance, obesity, diabetes, dyslipidemia, nonalcoholic fatty liver, and other disorders (8–11). There is growing evidence that the maternal nutritional status can alter the state of the fetal genome and imprint gene expression. Barker hypothesizes that the malnourished fetus is programmed to exhibit a “thrifty phenotype” with an increase in appetite and adipocyte activity, fat deposition, a decrease in glomerular and vascular compliance, a reduction in pancreatic beta cells and insulin signaling, and possibly decreased energy output (12,13), although the mechanisms responsible for fetal programming remain poorly understood. In some animal models, aspects of fetal programming can be reserved postnatally. Anatomic and functional (cell signaling) changes, such as a decrease in GLUT4 induction after insulin treatment, had been described in affected individuals.
To assess these potential epigenetic changes, we developed an IUGR model in the rat whereby the animals developed insulin resistance by between 3 and 6 mo of age. It was hypothesized that early nutrition and catch-up growth could have long-term effects on insulin resistance in IUGR. Here, we compared the gene expression and glucose transport in skeletal muscle cells in IUGR rats to normal rats and confirmed that the SOCSs were key suppressor genes, which could influence glucose transport leading to insulin resistance.
The SOCS (also known as JAB and SSI) proteins play an important role in the pathogenesis of metabolic syndrome by concordantly modulating cytokine and insulin signaling. An increase in SOCS is recognized as one of the important causes of insulin resistance. The SOCS family is composed of SOCS-1 to -7 and the cytokine-inducible src homology 2 domain-containing protein (CIS). These proteins are thought to participate in negative feedback loops in cytokine signaling. SOCS-1 and SOCS-3 bind JAK tyrosine kinase and attenuate its ability to phosphorylate signal transducer and activator of transcription (STAT) proteins. In addition, CIS and SOCS-3 bind phosphorylated cytokine receptors and competitively interfere with binding of other src homology 2 domain-containing proteins (14,15). Expression of SOCS proteins is increased by cytokine signaling through activation of STAT- and NF-κB-mediated pathways. Thus, the negative feedback loop via SOCS proteins is doubly regulated in both a phosphorylation-dependent manner and a transcription-dependent manner (16). Recent studies using a yeast two-hybrid system and molecular reconstitution in cultured cells have shown that SOCS-1, SOCS-3, and SOCS-6 bind insulin receptors and that SOCS-2 and SOCS-3 can bind the IGF 1 receptor. If SOCS proteins were able to attenuate insulin signaling in vivo, they would be attractive candidate molecules for linking increased cytokine levels and decreased insulin sensitivity in insulin-resistant states (17).
This study demonstrated that SOCS-1 and SOCS-3 increased, and IRS-1 decreased in IUGR-born adult rats under insulin-resistant states. The elongin BC-binding motif in SOCS-1 and SOCS-3 is required for the ubiquitination and degradation of IRS-1 and IRS-2, revealing an additional mechanism to inhibit insulin action and might promote long-term insulin resistance. It was previously reported that increased levels of SOCS-1 and SOCS-3 bind to distinct domains of insulin receptors, inhibiting phosphorylation of IRS-1 and IRS-2. Subsequent downstream signaling was also inhibited but without affecting tyrosine phosphorylation of IR leading to insulin resistance. This attenuation of insulin signaling resulted in a reduction of activation of glycogen synthesis and glucose transport in cultured cells (5). Moreover, increased levels of SOCS-1 and SOCS-3 could also cause a prominent up-regulation of the key regulator of fatty acid synthesis in liver and sterol regulatory element-binding protein (SREBP)-1. In contrast, inhibition of SOCS-1 and SOCS-3 in livers of obese diabetic db/db mice by antisense treatment modestly improves insulin sensitivity and completely normalizes the increased expression of SREBP-1 that are dramatic amelioration of hepatic steatosis and hypertriglyceridemia (6,18). This study confirmed that these two SOCS proteins have unique mechanisms of attenuation of insulin signaling, suggesting that both SOCS-1 and SOCS-3 could serve as therapeutic targets for type 2 diabetes and other insulin-resistant states.
RNAi is a procedure in which double-stranded RNA (dsRNA) triggers the degradation of a homologous mRNA. Compared with antisense technology, RNAi seems to be more efficient in silencing target gene expression with sequence and strand specificity. As a normal physiological regulation pathway, the RNAi pathway is important in regulating endogenous gene expression during an organism's development. Furthermore, because of its specificity, RNAi could also be used as a means of gene therapy. In the study described herein, four different shRNA sequences against SOCS-1 and -3 were designed and ultimately transfected into skeletal muscle cells. The recombinant plasmid significantly inhibited the expression of SOCS-1 and SOCS-3 at both the mRNA and protein levels.
GLUT is the main cellular transporters that mediated the uptake of glucose. GLUT4 is the major glucose transporter in muscle. A defect in insulin action leading to the plasma membrane incorporation of GLUT4 may result in impaired glucose transport (19–21). Decreased AKT phosphorylation affects the activity of GLUT4, which translocates glucose into cells. As a result, blood levels of glucose increase. The subcellular distribution, gene expression, and inherent activity of GLUT4 directly affect the transmembrane transportation of glucose induced by insulin (22). By strengthening the hexosamine pathway, high glucose levels and hyperinsulinemia can inhibit transposition of GLUT4. In turn, this results in a down-regulation of protein level and tyrosine phosphorylation of IRS and a depressed expression of GLUT4 at the transcriptional and posttranscriptional levels. The inherent activity of GLUT4 is directly reduced, GLUT4-induced glucose transportation decreased, and insulin resistance results (23).
Andersen et al. (24) previously reported that GLUT4 gene expression in muscle was acutely stimulated by short-term insulin stimulation in healthy subjects, but that GLUT4 mRNA expression failed to increase after insulin infusions in type 2 diabetic patients and insulin-resistant first-degree relatives. Together, these data support hypothesis that impaired insulin stimulation of GLUT4 expression would reflect insulin resistance itself. In this investigation, after transfecting the most effective shRNA recombinant plasmids into IUGR skeletal muscle cells, not only was a significant inhibition in SOCS-1 and SOCS-3 at both the mRNA and protein levels detected but also a promoting effect on GLUT4 translocation with insulin, with higher extend of Akt phosphorylation in response to insulin stimulation. Together, these findings suggested that SOCS-1 and SOCS-3 knockdown ameliorated GLUT4 translocation under insulin-stimulated conditions in IUGR individuals.
In conclusion, we successfully constructed a rat model of IUGR with insulin resistance and determined that the decreased expression of IRS-1 and increased expression of SOCS-1 and SOCS-3 occurred. Furthermore, this study provided evidence that malnutrition in utero is associated with changes in the expression of key molecules in insulin pathways in muscle; these changes may be related to insulin resistance in IUGR-born adults. Moreover, shRNA are capable of down-regulating the expression of SOCS-1 and SOCS-3 in IUGR skeletal muscle cells in vitro, simultaneously ameliorating insulin-stimulated glucose transport via increasing the activity of GLUT4. Together, these results imply that both SOCS-1 and SOCS-3 could serve as therapeutic targets for insulin-resistant states in IUGR-born adults.
Abbreviations
- GLUT:
-
glucose transporter
- HOMA-IR:
-
homeostasis model assessment of insulin resistance
- IRS-1:
-
insulin receptor substrate-1
- SOCS:
-
suppressors of cytokine signaling
- shRNA:
-
small hairpin RNAs
REFERENCES
Neitzke U, Harder T, Schellong K, Melchior K, Rodekamp E, Dudenhausen JW, Plagemann A 2008 Intrauterine growth restriction in a rodent model and developmental programming of the metabolic syndrome: a critical appraisal of the experimental evidence. Placenta 29: 246–254
Yajnik CS 2004 Early life origins of insulin resistance and type 2 diabetes in India and other Asian countries. J Nutr 134: 205–210
Grundy SM, Brewer BH, Cleeman JI, Smith SC, Lenfant C 2004 Definition of metabolic syndrome. Circulation 109: 433–438
Virkamäki A, Ueki K, Kahn CR 1999 Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J Clin Invest 103: 931–943
Ueki K, Kondo T, Kahn CR 2004 Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol Cell Biol 24: 5434–5446
Starr R, Metcalf D, Elefanty AG, Brysha M, Willson TA, Nicola NA, Hilton DJ, Alexander WS 1998 Liver degeneration and lymphoid deficiencies in mice lacking suppressor of cytokine signaling-1. Proc Natl Acad Sci U S A 95: 14395–14399
Shahkhalili Y, Moulin J, Zbinden I, Aprikian O, Mace K 2010 Comparison of two models of intrauterine growth restriction for early catch-up growth and later development of glucose intolerance and obesity in rats. Am J Physiol Regul Integr Comp Physiol 298: R141–R146
Frontini MG, Srinivasan SR, Xu J, Berenson GS 2004 Low birth weight and longitudinal trends of cardiovascular risk factor variables from childhood to adolescence:the bogalusa heart study. BMC Pediatr 4: 22–25
Lucas A 2005 Long-term programming effects of early nutrition—implications for the preterm infants. J Perinatol 25: S2–S6
Lebovitz HE 2006 Insulin resistance—a common link between type 2 diabetes and cardiovascular disease. Diabetes Obes Metab 8: 237–249
Ozanne SE, Fermandez TD, Hales CN 2004 Fetal growth and adult disease. Semin Perinatol 28: 81–87
Hales CN, Barker DJ 1992 Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 35: 595–601
Barker DJ, Eriksson JG, Forsen T, Osmond C 2002 Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol 31: 1235–1239
Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimuna S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A 1997 A new protein containing an SH2 domain that inhibits JAK kinases. Nature 387: 921–924
Peraldi P, Filloux C, Emanuelli B, Hilton DJ, Van Obberghen E 2001 Insulin induces suppressor of cytokine signaling-3 tyrosine phosphorylation through janus-activated kinase. J Biol Chem 276: 24614–24620
Ain R, Canham LN, Soares MJ 2005 Dexamethasone-induced intrauterine growth restriction impacts the placental prolactin family, insulin-like growth factor-II and the Akt signaling pathway. J Endocrinol 185: 253–263
Mooney RA, Senn J, Cameron S, Inamdar N, Boivin LM, Shang Y, Furlanetto RW 2001 Suppressors of cytokine signaling-1 and -6 associate with and inhibit the insulin receptor, a potential mechanism for cytokine-mediated insulin resistance. J Biol Chem 276: 25889–25893
Ueki K, Kadowaki T, Kahn CR 2005 Role of suppressors of cytokine signaling SOCS-1 and SOCS-3 in hepatic steatosis and the metabolic syndrome. Hepatol Res 33: 185–192
Garvey WT, Huecksteadt TP, Matthakt S, Olefsky JM 1988 Role of glucose transporters in the cellular insulin resistance of type 2 non-insulin-dependent diabetes mellitus. J Clin Invest 81: 1528–1536
Jaquet D, Vidal H, Hankard R, Czernichow P, Levy-Marchal C 2001 Impaired regulation of transporter 4 gene expression in insulin resistance associated with in utero undernutrition. J Clin Endocrinol Metab 86: 3266–3271
Carvalho E, Schellhorn SE, Zabolotny JM 2004 GLUT4 overexpression or deficiency in adipocytes of transgenic mice alters the composition of GLUT4 vesicles and the subcellular localization of GLUT4 and IRAP. J Biol Chem 279: 21598–21605
Thamotharan M, Shin BC, Suddirikku DT 2005 GLUT4 expression and subcellular localization in the intrauterine growth-restricted adult rat female offspring. Am J Physiol Endocrinol Metab 288: E935–E947
Holness MJ 2001 Enhanced glucose uptake into adipose tissue induced by early growth restriction augments excursions in plasma leptin response evoked by changes in insulin status. Int J Obes Relat Metab Disord 25: 1775–1781
Andersen PH, Lund S, Vestergaard H, Junker S, Kahn BB, Pedersen O 1993 Expression of the major insulin regulatable glucose transporter (GLUT 4) in skeletal muscle of non insulin dependent diabetic patients and healthy subjects before and after insulin infusion. J Clin Endocrinol Metab 77: 27–32
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Natural Science Foundation of China (No.30672262 and No.30772358) [X.L.], Eleventh Five-year National Science Supported Planning Project (No.2006BAI05A07) [X.L.], the Doctor Program of Higher Education from the Ministry of Education of China (No. 20060487062) [X.L.], and the 973 National Basic Research Project from the Ministry of Science and Technology of China (No. 2007CB512900) [Q.N.].
Rights and permissions
About this article
Cite this article
Liao, L., Zheng, R., Wang, C. et al. The Influence of Down-Regulation of Suppressor of Cellular Signaling Proteins by RNAi on Glucose Transport of Intrauterine Growth Retardation Rats. Pediatr Res 69, 497–503 (2011). https://doi.org/10.1203/PDR.0b013e31821769bd
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e31821769bd
This article is cited by
-
LRP6 Bidirectionally Regulates Insulin Sensitivity through Insulin Receptor and S6K Signaling in Rats with CG-IUGR
Current Medical Science (2023)
-
Withania somnifera as a potential candidate to ameliorate high fat diet-induced anxiety and neuroinflammation
Journal of Neuroinflammation (2017)
-
IUGR with infantile overnutrition programs an insulin-resistant phenotype through DNA methylation of peroxisome proliferator–activated receptor-γ coactivator-1α in rats
Pediatric Research (2015)
-
Polymorphisms in the SOCS7 gene and glucose homeostasis traits
BMC Research Notes (2013)
-
Effects of SOCS 1/3 gene silencing on the expression of C/EBPα and PPARγ during differentiation and maturation of rat preadipocytes
Pediatric Research (2013)