Abstract
Stem cell-like brain tumor initiating cells (BTICs) cause recurrence of glioblastomas, with BTIC ‘stemness’ affected by epigenetic mechanisms. The ING family of epigenetic regulators (ING1-5) function by targeting histone acetyltransferase (HAT) or histone deacetylase complexes to the H3K4me3 mark to alter histone acetylation and subsequently, gene expression. Here we find that ectopic expression of ING5, the targeting subunit of HBO1, MOZ and MORF HAT complexes increases expression of the Oct4, Olig2 and Nestin stem cell markers, promotes self-renewal, prevents lineage differentiation and increases stem cell pools in BTIC populations. This activity requires the plant homeodomain region of ING5 that interacts specifically with the H3K4me3 mark. ING5 also enhances PI3K/AKT and MEK/ERK activity to sustain self-renewal of BTICs over serial passage of stem cell-like spheres. ING5 exerts these effects by activating transcription of calcium channel and follicle stimulating hormone pathway genes. In silico analyses of The Cancer Genome Atlas data suggest that ING5 is a positive regulator of BTIC stemness, whose expression negatively correlates with patient prognosis, especially in the Proneural and Classical subtypes, and in tumors with low SOX2 expression. These data suggest that altering histone acetylation status and signaling pathways induced by ING5 may provide useful clinical strategies to target tumor resistance and recurrence in glioblastoma.
Similar content being viewed by others
Introduction
Glioblastoma multiforme (GBM) is the most frequent and lethal type of intracranial tumor with a five-year survival rate of 5.1%.1, 2 An intratumoral lineage hierarchy has been uncovered following the identification of tumor initiating cells in brain tumors,3, 4 and the progenitor-like status of brain tumor initiating cells (BTICs) is thought to make gliomas particularly refractory to standard cancer treatments. Primary cultures of BTICs can establish tumors during serial in vivo transplantation5, 6 and repopulate tumor sites after irradiation and chemotherapy.7, 8 BTICs display a combination of neural stem cell phenotypes and tumorigenic properties, and may originate from crosstalk between deregulated developmental processes and mitogenic mutations during gliomagenesis.9 Both neurodevelopmental factors and mitogenic signaling pathways contribute to the ‘stemness character’ of BTICs. For example, the Notch and inhibitor of differentiation (ID) proteins promote self-renewal in both neural stem cells and BTICs.10, 11, 12 EGFR and PDGFR activation, the most common genetic aberrations in gliomas, can induce de-differentiation of mature astrocytes to cause gliomagenesis in mouse models,13, 14 and promote BTIC maintenance in vitro.15
Epigenetic alterations drive oncogenesis16 and in GBM, there is a disproportionately high rate of mutation in chromatin modifiers.17 The epigenetic landscape, which is distinct between BTICs and differentiated GBM cells, helps maintain BTIC identity and prevents interconversion between these cell types.18, 19 This epigenetic barrier between BTICs and non-stem GBM cells can be traversed via epigenetic remodeling that resembles reprogramming of induced pluripotent stem cells.20 However, there are limited studies about specific epigenetic factors in BTIC regulation, with most focusing on members of the polycomb group and mixed-lineage leukemia.21, 22
The inhibitor of growth (ING) family of epigenetic regulators (ING1-5) affect growth, the DNA damage response, cellular senescence and apoptosis.23, 24, 25, 26 INGs are stoichiometric components of histone acetyltransferase (HAT) or histone deacetylase (HDAC) complexes27, 28, 29 and target them specifically to the H3K4me3 mark with their plant homeodomain (PHD) form of zinc finger.30 Although initially identified as type II tumor suppressors and intensively studied in cancer systems,31 INGs also function in germ cell development and cell differentiation via epigenetic regulation.32, 33, 34
ING5 has been reported to regulate cell proliferation,35, 36 apoptosis,37 metastasis38 and chemoresistance37, 39 in different cancer models. Like other ING family members, ING5 can bind and acetylate p53 by associated HAT complexes,40, 41 which is assumed to account for most of its inhibitiory effects on proliferation seen in different cancers.37, 42, 43, 44 However, ING5 also possesses p53-independent functions in promoting the proliferation of several cancer cell lines, probably due to interactions with HBO128 and CDK2.35 ING5 is rarely mutated in cancer but the deregulation of its expression has been reported in many cancer types, with decreased expression in lung cancer and acute myeloid leukemia36, 44 and increased expression in breast cancer and glioblastoma.42, 43 Overall, the role of ING5 in cancer appears to be highly tissue specific and a discrepancy between its nuclear and cytoplasmic levels noted in previous studies complicates the investigation of its functions. ING5 appears to be more multifunctional than other ING proteins, being incorporated into the HBO1, MOZ and MORF HAT complexes.28, 45 ING5 serves as a critical epigenetic regulator for epidermal stem cell maintenance with global co-occupancy with H3K4me3 at genes actively expressed in stem cells and silenced during differentiation.46 High ING5 levels were also seen in pluripotent cells compared to differentiated lineages,47, 48, 49 suggesting it may function as a common factor for cell plasticity in stem cell systems.
Since ING5–HAT complexes were reported to function in the maintenance of pluripotent cells (HBO1),50 hematopoietic stem cells (MOZ)51 and neural stem cells (MORF),52 we asked whether ING5 affected stem cell properties of BTICs and indeed, found that it does. ING5 is highly expressed in stem cells where it promotes BTIC self-renewal most strikingly in the absence of growth factors. Pathway analyses indicate ING5 promotes stemness by maintaining intracellular calcium levels and activating the follicle stimulating hormone (FSH) pathway, which co-operatively contribute to BTIC self-renewal, providing novel candidates for targeting BTICs in GBMs.
Results
ING5 maintains BTIC self-renewal
E14, R1 and D3 mouse embryonic stem cell lines showed an immediate reduction of ING5 mRNA (Supplementary Figure S1a) and protein (Supplementary Figures S1b, c) as pluripotent cells differentiated. The intrinsic stem cell properties of primary BTIC cultures are best preserved in serum-free medium supplemented with growth factors in which they grow as spheres,15 and upon 1% serum treatment they differentiate into multiple neural lineages identified by cell type-specific markers (Figure 1a). In the BT 189 cell strain we also observed decreased ING5 mRNA and protein within the first two days of serum addition (Figures 1b–d). ING5 immunofluorescence was weak nuclear localization was lost in differentiated cells (Figure 1b). Two additional cell strains, BT 12 and BT 134 also showed rapid decline of ING5 upon differentiation (Figure 1e). Sphere formation assays of cells seeded at low density showed that pCI-ING5 transfection increased both the proportion of clonogenic cells (sphere formation rate) and the size of spheres (Figure 1f). Conversely, ING5 downregulation by siRNA reduced formation and size of spheres (Figure 1g). To achieve homogeneous knockdown, we constructed lentiviral-based shR-ING5 strains (shR1, shR2) with RFP labels. Two independent knockdown strains showed reduced sphere-forming capability (Figure 1h) and the shR2 strain (hereafter shR-ING5) with an 80% knockdown efficiency was used in subsequent experiments. The ING5-HBO1 HAT complex was also reported to function in the initiation of DNA replication.28 To determine if increased sphere-forming rates were due to cell proliferation, we analyzed cell cycle profiles of BTICs after ING5 overexpression and knockdown, but did not observe an effect of ING5 on cell cycle distribution, suggesting that higher sphere-forming ability reflected an increase in asynchronous self renewal of cells (Supplementary Figure S1d). The expression levels of stem cell core transcription factors OCT4 and OLIG2, and neural stem cell markers CD133 and Nestin were upregulated several-fold by ING5 (Figure 1i). Induction of Nestin and repression of neuronal lineage marker Tubulin β III (Tubb3) was also seen by western blot in ING5 overexpressing sphere cells (Figure 1j) and reciprocal regulation was seen in ING5 knockdown cells (Figure 1k).
ING5 maintains BTIC self-renewal. (a) Morphology of self-renewing spheres and immunofluorescence of neuronal and glial lineage markers in differentiated cells. Scale bar=200 μm. (b) Immunofluorescence of ING5 in undifferentiated (upper panels) and cells differentiated for 5 days (lower panels). Scale bar=20 μm. (c, d) The mRNA levels (c) and protein levels (d) of ING5 decrease during differentiation in BT 189 cells. n=4. (e) Immunoblotting of ING5 protein in the BT 12, and BT 134 lines during differentiation. (f) ING5 overexpression increases sphere formation rates and average volumes in BTIC sphere formation assays (n=3, **P<0.01, *P<0.05). Scale bar=400 μm. (g) ING5 knockdown by siRNA decreases sphere formation rates and sphere sizes of BTICs (n=4, **P<0.01, *P<0.05). Scale bars=400 μm. (h) (Left) Sphere formation rates for cell lines stably expressing shRNAs against ING5 (shR1 and shR2-ING5) or control non-targeting shRNA (shR-ctr). (n=4, **P<0.01, *P<0.05). (Right) Fluorescence of the RFP reporter in stable cell lines superimposed with differential interference contrast (DIC) images. Scale bar=100 μm. (i) RT-qPCR analysis of stem cell core transcription factors and stem cell markers after ING5 overexpression. (j, k) Western blot analysis of the neural stem cell marker Nestin and neuronal lineage marker Tubb3 in response to ING5 overexpression (j) and knockdown (k).
ING5 increases the stem cell pool and inhibits differentiation
BTICs grow as heterogeneous populations in vitro with a spectrum of variable self-renewal capabilities. To characterize stem cell hierarchy in BTICs we utilized the CD44 and CD133 cell surface markers, which strongly correlate with the stemness properties and tumor initiating abilities of glioma cells.53, 54 We used a transposon-based PiggyBac system (iPB) for stable and inducible ING5 overexpression.55 iPB-ING5 cells treated by the inducer cumate showed a higher percentage of CD44 and C133 positive cells compared to the control (iPB-ctr) (Figure 2a, upper panels). Conversely, shR-ING5 reduced CD44 positive cells (Figure 2a, lower panels), indicating that ING5 increased the stem cell pool in BTICs.
ING5 increases the stem cell pool and inhibits differentiation. (a) Flow cytometry analysis of CD133/CD44 positive cells in iPB cell lines (top panels) and CD44 positive cells in shRNA cell lines (bottom panels), gated by isotype control. (b) (Left) Mitotic pair analysis of the three division modes: symmetric proliferating (sym-pro), symmetric differentiating (sym-diff) and asymmetric (asym) cell division, in iPB cell lines. Over 150 pairs were counted for each group in one experiment. n=3, * P<0.05. (Right) An example of cell division symmetry based on the distribution of stem cell factor Nestin with the Red arrow indicating symmetric differentiating division and the white arrow asymmetric division. (c) Morphological changes of iPB cells before (Day 0) and after (Days 1–3) differentiation induced by 1% FBS. Scale bar=100 μm. (d) After differentiation for 5 days, immunofluorescence of Nestin and Tubb3 in shRNA cell lines with an RFP reporter. Scale bar=200 μm. (e) Western blot of Nestin and Tubb3 in differentiated shRNA cell lines corresponding to cells shown in d.
Self-renewal is achieved in tissue stem cells through uneven distribution of fate determinants for stemness via asymmetric cell division.56 In neural stem cells, CD133 specifically marks the apical membrane initiating subsequent asymmetric division.57 Since ING5 reciprocally altered CD133 levels, we tested if ING5 also altered symmetry of cell division. In our mitotic pair analysis, the distribution of Nestin was used as an indicator for three division modes, symmetric proliferating, symmetric differentiating and asymmetric division (Figure 2b). Despite a high proportion of symmetric division probably due to perturbed asymmetric machinery in tumor initiating cells,58 ING5 induced a more than two-fold increase of asymmetric divisions (highlighted by the white arrowhead in Figure 2b), indicating a function in the intrinsic self-renewal machinery of this BTIC strain.
Ectopic expression of ING5 also interfered with lineage commitment. iPB-ING5 cells slowed FBS-induced morphological changes compared to control cells (Figure 2c) and immunostaining indicated a higher level of the neural stem cell marker Nestin and reduced level of Tubb3 in iPB-ING5 cells (Supplementary Figures S2a, b). Conversely, shR-ING5 knockdown cells displayed higher levels of Tubb3 and less Nestin (Figure 2d), which was corroborated by western blotting (Figure 2e). These data suggest ING5 inhibits lineage commitment in BTICs.
Enhanced mitogenic signaling by ING5 promotes long-term self-renewal
We next performed a more stringent sphere formation assay in the absence of growth factors. EGF and FGF are required to support propagation of neural stem cells and BTICs in vitro.59, 60 Sphere formation was substantially impaired without EGF and FGF, but under these conditions ING5 more strongly promoted sphere formation (Figure 3a). ING5 maintained long-term self-renewal over serial sphere passages, strongly contrasting with the complete exhaustion of stem cells seen in the control group (Figures 3a and b and Supplementary Figures S2c, d). To test if this correlated with mitogenic signaling, we used the kinase inhibitors Tofacitinib, PX-866 and PD184352 that target the JAK/STAT, PI3K/Akt and MEK pathways, respectively. Tofacitinib impaired sphere formation in both control and ING5 overexpressing groups (Figure 3c). However, ING5 overexpressing cells displayed resistance to the MEK1/2 and PI3K inhibitors and retained sphere-forming ability compared to controls (Figure 3c). Consistent with this, ING5 increased levels of phosphorylated AKT and ERK1/2 (Figure 3d) and inhibiting either PI3K or MEK1/2 induced the neuronal lineage marker Tubb3 and suppressed Nestin in both iPB-ING5 and iPB-ctr cells (Figure 3e). We also found that inhibition of the MEK/ERK pathway, but not PI3K/AKT signaling, reduced the CD133 positive pool in the BTIC population from 76 to 52% (Figure 3f). Therefore, the MEK/ERK pathway appears to be more directly related to ING5-mediated self-renewal of BTICs than the other pathway tested.
ING5 activates mitogenic pathways to promote self-renewal. (a) The sphere formation rates of iPB-ctr and iPB-ING5 overexpressing cells at three successive passages in the absence of EGF and FGF treatment. (n=3, **P<0.01, ***P<0.001). (b) DIC images of spheres from the tertiary sphere passage in iPB cells. Scale bar=400 μm. (c) Sphere formation rate under treatment with protein kinase inhibitors (n=3, *P<0.05). (d) Protein and phosphorylated protein levels of effectors in the PI3K and MEK pathways. Cells were treated with PX-866 at 1 μM and PD184352 at 2 μM for 48 h. (e) Immunofluorescence of Nestin and Tubb3 in differentiated iPB control (Left panels) or iPB-ING5 cells (Right panels) treated with 1 μM PX-866 or 2 μM PD184352. Scale bar=200 μm. (f) Flow cytometry analysis of the CD133 positive population in PX-866 (1 μM) and PD184352 (2 μM) treated BTIC 189 cells, gated by isotype control.
Calcium (Ca2+) channel activity is induced by ING5
Transcriptome assays using ING5 knockdown cells (shR-ctr and shR-ING5) (Supplementary Table S1; GSE100398) were hierarchically clustered and showed consistency among replicates (Figure 4a). Most differentially expressed genes were silenced by ING5 knockdown, suggesting a primary function of ING5 in gene activation. Gene set enrichment analysis (GSEA) identified Ca2+ channel activity as the top ranked Gene Ontology (GO) term and four additional GO terms related to cation channels were among the top 25 (Figure 4b). Ca2+ channel components upregulated by ING5 (Figure 4c) include the subunits of L-type (CACNA1F, CACNA1S, CACNA1D and CACNA1C) and P/Q-type (CACNA1A) voltage-gated Ca2+ channels, and transient receptor potential cation channels permeable to Ca2+ (TRPC3, TRPC5, TRPC4, TRPM1). A subset of these genes was validated using real-time quantitative PCR (qPCR) (Figure 4d), with most of the genes showing reciprocal effects between ING5 knockdown and overexpression (Supplementary Figure S3a). GO analysis also generated a high ranking for the category including the neurotrophic tyrosine receptor kinases NTRK1 and NTRK2, which was corroborated using qPCR (Figures 4e and f). These NTRKs can activate MAPK in the absence of EGF61 and might contribute to mitogenic signaling induced by ING5. Reduction of the neurotrophic pathway during differentiation was observed in parallel with ING5 downregulation (Supplementary Figure S3b). Chromatin immunoprecipitation (ChIP) assays showed ING5 binding the NTRK1 promoter correlated well with NTRK1 transcriptional induction (Supplementary Figure S3c), specifically of the variants including an exon highly expressed in the human brain.62
Transcriptome analysis indicates ING5 activates calcium channels. (a) Hierarchical clustering of microarray data in four samples, showing two clusters (shR-NT vs shR-ING5) based on ING5 status. The color scale is in log base 2. (b) Top 25 hits using GSEA GO analysis. NES, normalized enrichment score. (c) Gene list and enrichment plot showing enrichment of genes upregulated in the NT group within the GO term calcium channel activity, with a positive correlation. Black bars indicate the position of genes in the ranked list of differentially expressed genes between NT and shR-ING5. The green line represents the enrichment score. (d) Validation of ING5 induced expression of genes encoding calcium channel components using real-time qPCR (n=3, *P<0.05, **P<0.01). (e) Enrichment plot and list of genes enriched in the GO term growth factor binding. (f) RT-qPCR shows the expression of genes in the neurotrophic pathways and KSR2 from the microarray data are elevated by ING5.
ING5-induced intracellular Ca2+ elevation is associated with self-renewal
Live cell imaging of Ca2+ using the fluorescent calcium dye Fluo3-AM showed that intracellular Ca2+ levels were increased by ING5 overexpression in iPB-ING5 cells (Figure 5a) and strongly reduced in shR-ING5 cells (Figure 5b). To examine ING5 and Ca2+ levels in individual cells, we co-transfected pCI-ING5 or pCI vector with pCI-mCherry and measured Fluo3-AM fluoresence intensity in mCherry positive cells. pCI-ING5 transfected cells showed elevated Ca2+ compared to controls (Supplementary Figure S4a). A reduction of the Ca2+-high population was seen consistently in shR-ING5 cells, while in iPB-ING5 cells a shift to the Ca2+-high population was seen (Figures 5c and d).
ING5 maintains high intracellular calcium levels to promote BTIC self-renewal. (a, b) Live cell calcium imaging in iPB (left panels) and shRNA (right panels) cell lines. Fluo3-AM (7 μM) was loaded for 1 h before imaging. Scale bar=400 μm. (c, d) Flow cytometry analysis of Fluo3-AM fluorescence intensity in shRNA (c) and iPB (d) cell populations, gated by calcium chelator BAPTA-AM treatment control. (e) Sphere formation rate in shRNA cell lines treated with Ionomycin, CPA, BAPTA-AM, Nifedipine or SKF96365 at the indicated concentrations for 12 days (n=3, **P<0.01 compared to shR-ctr/DMSO, #P<0.05 and ##P<0.01 compared to shR-ING5/DMSO). (f) Flow cytometry analysis of CD133 positive cells in BTIC 189 cells treated with calcium inducers Ionomycin (20 nM) or CPA (100 nM) for 2 days, gated by isotype control. (g) The effect of Ionomycin treatment on levels of phosphorylated AKT and ERK1/2 in the PI3K and MEK pathways, respectively. Cells were treated with 10 nM Ionomycin for 48 h. (h) Sphere formation rate in iPB cells treated with KN-93 or Cyclosporin A at the indicated concentrations. n=3, *P<0.05 compared to iPB-ctr/DMSO and #P<0.05 compared to iPB-ING5/DMSO.
Ca2+ signaling positively correlates with GBM cell survival63, 64 and promotes stem cell features in lung cancer cells.65 To ask if Ca2+ levels correlated with BTIC stem cell properties, we tested the effects of Ca2+ modulators on single cell sphere formation. Ionomycin and cyclopiazonic acid (CPA) increase intracellular Ca2+ through plasma membrane import and store-operated entry, respectively (Supplementary Figure S4b), and they both promoted sphere-forming capabilities, with the rates of sphere formation in knockdown cells nearing the level seen in the untreated (DMSO) shR-ctr group (Figure 5e). By contrast, treatment with the Ca2+ chelator BAPTA-AM, or the L-type (Nifedipine) and T-type (SKF96365) Ca2+ channel blockers reduced intracellular Ca2+ levels and decreased sphere-forming capabilities (Figure 5e). Ionomycin and CPA treatment also increased the pool of CD133 positive BTICs (Figure 5f). The above reagents did not induce cell apoptosis at the concentrations used, as estimated by cell morphology and PARP cleavage (Supplementary Figure S4c).
Since Ca2+ signaling is closely associated with PI3K and MAPK signaling pathways through downstream effectors PKC, calcineurin (CaN) and calmodulin-dependent kinases (CAMKs), we tested whether increasing Ca2+ affected these pathways in BTICs. We found both phosphorylated AKT and ERK were elevated by Ionomycin treatment (Figure 5g), suggesting that the robust activation of PI3K and MEK/ERK signaling by ING5 (Figure 3d) occurred at least in part, through Ca2+ signaling. We further treated cells with the CAMKII inhibitor KN-93 and the CaN inhibitor Cyclosporin A (CsA) and found that CsA, but not KN-93 treatment abolished the self-renewal-promoting effects of ING5 in iPB-ING5 cells (Figure 5h), suggesting that Ca2+ signaling was mainly transduced through CaN. The phosphatase CaN can activate the MEK/ERK pathway through its substrate Kinase Suppressor of Ras 2 (KSR2) that promotes Raf-mediated MEK phosphorylation.66 KSR2 increased 9-fold in response to ING5 in the microarray, and was validated by qPCR (Figure 4f).
The FSH pathway mediates ING5 function in parallel with Ca2+ signaling
Ionomycin and CPA treatment increased sphere formation in iPB-ctr cells, but not to levels seen in iPB-ING5 cells (Figure 6a), indicating that Ca2+ did not transduce all of the effects of ING5. IPA pathway analysis yielded a high z-score for the FSH pathway (Figure 6b), which also appeared in the top 25 GO term ‘Female Gamete Generation’ in Figure 4b. Both the hormone (FSHB) and the receptor (FSHR) were induced by ING5 in the microarray and this was confirmed by qPCR (Figure 6c). Expression of both genes also decreased during differentiation (Figure 6d).
The FSH pathway transduces effects of ING5 on stem cell properties. (a) Sphere formation assay for cells treated with calcium modulators and FSHR blocking antibody (Anti-FSHR) at the indicated concentrations (n=3, *P<0.05 and **P<0.01 compared to iPB-ctr/DMSO; #P<0.05 and ##P<0.01 compared to iPB-ING5/DMSO). (b) IPA downstream function analysis indicates the FSH pathway is elevated by ING5. Genes positively correlated with this function were listed with fold changes. (c) RT-qPCR of genes related to hormone and steroidogenesis functions. (n=3, *P<0.05, **P<0.01) (d) The expression levels of FSHB and FSHR genes in BT 189 cells before and after differentiation for 1–5 days. (e) Immunostaining for ING5 and FSHR in iPB cells. Scale bar=100 μm. (f) Flow cytometry analysis of CD133 positive cells in BT 189 cells treated with FSHR neutralizing antibody or IgG control, gated by isotype control. (g) Immunofluorescence of Nestin and Tubb3 shows inhibition of the FSH pathway induces neuronal differentiation. Scale bar=200 μm. (h) FSH recombinant protein treatment at indicated concentrations increases sphere formation rates in shRNA cell lines (n=3, *P<0.05 and **P<0.01 compared to untreated shR-ctr; #P<0.05 compared to untreated shR-ING cells). (i) FSH recombinant protein treatment induces sphere-forming abilities in iPB-ctr cells but not in ING5 overexpressing cells (n=3, **P<0.01). (j) Sphere formation rates for cells treated with Anti-FSHR or BAPTA alone, and the combination of both (n=3, *P<0.05). (k) FSH treatment at 5 ng/ml for 3 days induces the expression of OCT4 and Nestin in BT 189 cells.
FSHR can activate the MAPK pathway67 and induces a stem cell-like phenotype through the ERK–OCT4 axis in ovarian cancer cells.68, 69 The FSH pathway plays essential roles in follicular development and germ cell generation,70 while its function in brain cells remains unexplored. The Cancer Genome Atlas (TCGA) data showed modest expression levels of FSHR and FSHB in GBM, but we found that ING5 strongly induced the level of FSH receptors on the plasma membrane (Figure 6e). To test if this pathway affected GBMs, we treated cells with an FSHR neutralizing antibody (anti-FSHR) that blocks ligand–receptor binding. This compromised sphere-forming ability in iPB cells to a degree comparable with the calcium inhibitors (Figure 6a). Anti-FSHR treatment also decreased the CD133 positive population (Figure 6f) and promoted neuronal differentiation (Figure 6g) compared to the IgG isotype control. Adding recombinant FSH increased sphere-forming ability in shRNA cell lines and iPB-ctr cells, but not in the iPB-ING5 group, probably because the activation of the FSH pathway had reached a saturation point (Figures 6h and i).
Although FSH increases intracellular calcium in ovarian granulosa cells,71 we did not observe changes in calcium levels by anti-FSHR treatment. Treating cells concomitantly with FSHR neutralizing antibody and BAPTA caused an additive effect in inhibiting self-renewal (Figure 6j), indicating that the two pathways induced by ING5 primarily function independently to promote self-renewal. OCT4 and Nestin expression was also increased by recombinant FSH (Figure 6k), suggesting that induction of OCT4 may be a major effector of the FSH pathway in promoting BTIC stem cell properties.
The PHD motif of ING5 is required for maintaining BTIC stemness
Since the PHD of ING proteins interacts specifically with H3K4Me3 to target HAT and HDAC complexes to remodel chromatin,27, 72 we asked if an intact PHD was needed for iING5 function in BTIC self-renewal. Sphere formation ability of iPB cells overexpressing ING5 lacking the PHD (ΔPHD) was indistinguishable from the negative control (Figures 7a and b). ChIP assays in three cell strains; BT 189 with endogenous ING5, BT 189 cells overexpressing wildtype ING5 with a C-terminal Flag tag (iPB-ING5) and BT 189 cells overexpressing PHD-deleted ING5 with a Flag tag (iPB-ΔPHD) (Figure 7b), showed an enrichment of endogenous and overexpressed wildtype ING5 binding to the proximal promoter regions (–250, –50) of target genes (FSHR, FSHB, TRPM3, CACNA1D and OCT4, Figure 7c, Supplementary Figure S5a), suggesting a specific binding activity and consistent with the position of the H3K4me3 mark in these genes. By contrast, the ING5-ΔPHD protein immunoprecipitated with a Flag antibody (therefore excluding endogenous ING5) was not enriched in these promoter regions compared to the IgG control. We consistently found that the target genes of ING5 could not be induced by ectopic ING5-ΔPHD overexpression (Supplementary Figure S5b).
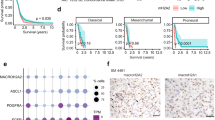
PHD motif is required for the function of ING5 in BTICs and ING5 levels negatively correlate with survival of GBM. (a) Sphere formation assays in iPB cell lines overexpressing wild-type (ING5-FLAG) and PHD-deleted ING5 (ΔPHD) (n=3, **P<0.01). (b) Western blot shows the protein levels of endogeneous ING5, wildtype ING5 with a FLAG tag and PHD-deleted ING5 (black arrows) in three iPB cell lines. (c) ChIP analysis of ING5 binding to promoters of target genes presented as fold enrichment relative to IgG controls. The endogenous ING5 in BT 189 cells, overexpressed ING5 with a Flag tag in iPB-ING5 cells and overexpressed PHD-deleted ING5 protein with a Flag tag were immunoprecipitated by the ING5 antibody and Flag antibody respectively. The upper panels are the schematic representation of the location of the primer sets and promoter regions enriched for ING5 binding were shown in red. (d) Kaplan–Meier survival analysis of TCGA GBM patients with high and low levels of ING5 expression (stratified by mean value, n=114). (e, f) ING5 expression levels negatively correlate with survival of the Proneural subtype (n=24) and the Classical subtype (n=30) of GBM patients. (g) The relationship of ING5 levels to survival in the SOX2-low group of patients (ING5, SOX2 stratified by median values, n=61). (h) Model for how ING5 functions in the maintenance of BTIC self-renewal. In the absence of growth factors, ING5 induces FSH and calcium signaling by promoting transcription of the FSH receptor and ligand genes, and various plasma membrane calcium channel genes. The FSH and calcium signaling pathways further activate PI3K/AKT and MEK/ERK signaling to induce stem cell features and the expression of stemness factors OCT4, OLIG2 and Nestin. Gene activation by ING5 is dependent on its PHD motif to target ING5-associated histone acetyltransferase complexes to the promoters.
High ING5 levels correlate with lower survival in a subset of GBMs
Kaplan–Meier analysis of TCGA data with patients divided into high or low ING5 groups showed a trend of lower survival associated with high ING5 expression (Figure 7d). Since GBMs are heterogeneous we analyzed four GBM subtypes (Proneural, Mesenchymal, Classical and Neural) separately and noted high ING5 levels correlated strongly with poor survival in the Proneural and Classical (Figures 7e and f) but not other (Supplementary Figure S5c) subtypes. It has been reported that BTIC lines exhibiting typical stem cell properties, but not progenitor features align with a Proneural signature.73 Single cell RNA-seq revealed that stemness signature was strongest in Proneural and Classical subtypes,74 suggesting that ING5 may be more relevant to the prognosis of tumors driven by the properties of stem cells.
Gene expression in 169 GBM samples revealed a linear relationship between the expression of SOX2 and RAF1 with ING5 (Supplementary Figure S5d; Supplementary Table S2). SOX2 is a master regulator and marker for stemness in neural stem cells.75 We stratified ING5 and SOX2 by median value, and a clear correlation between ING5 and SOX2 levels was shown in the crosstabulation (Supplementary Figure S5e) with the impact of ING5 on survival strong in the SOX2-Low (Figure 7g) but not the SOX2-High (Supplementary Figure S5f) group.
Discussion
To examine the role of ING5 in GBM we conducted sphere formation assays, CD44/CD133 profiling, asymmetric division analyses and differentiation assays, all of which indicated that ING5 functions to promote stemness of BTICs. Growth factor-receptor tyrosine kinase (RTK) signaling pathways are essential to preserve BTIC stem cell properties in vitro and we found that ING5 promoted self-renewal for several serial passages in the absence of growth factors, correlating closely with activation of the PI3K and MEK/ERK pathways. Transcriptome analyses also indicated that ING5 promotes mitogenic signaling through RAF1, NTRK receptors and calcium signaling. The PI3K and RAS/ERK pathways have been reported to block differentiation and induce dedifferentiation in mature neural lineages during gliomagenesis.76 Our results suggest that both the ERK and PI3K pathways inhibit neuronal differentiation in BTICs and the MEK/ERK pathway could also affect the CD133 positive stem cell pool. ERK signaling is known to antagonize neuronal differentiation by promoting OLIG2 expression77 and ING5 induces the expression of OLIG2, possibly mediated by the MEK/ERK pathway.
The PHD of ING5, which facilitates the targeting of HAT complexes to gene promoters containing the H3K4Me3 mark,45 is required for ING5's stemness-promoting function and binding to the proximal promoter regions of target genes. Consistent with the generally positive effects of HATs on gene expression, our transcriptome analysis indicates ING5 mainly functions through gene activation, representing a distinct, and possibly complementary mechanism from the well-known stemness factor PRC2 in BTIC maintenance by silencing the BMP4-induced differentiation pathway.78
In GBM, a number of RTKs and GPCRs activated by extracellular signals can potentially mobilize the second messenger Ca2+, however such a role of Ca2+ in BTIC regulation was previously unknown. We found that ING5 maintained relatively high levels by increased activity of membrane channels, and that such Ca2+ concentrations are required for self-renewal of BTICs. Calcium signaling is also closely associated with mitogenic pathways and we observed enhanced phosphorylation of AKT and ERK by increasing intracellular Ca2+ levels. We further found that CaN was a critical effector of Ca2+ signals that could potentially connect to the MEK pathway through its substrate KSR2.
Glycoproteins and steroid hormones have been reported to regulate different aspects of neurodevelopment and neural stem cell properties, where they usually play neurotrophic roles.79, 80, 81 We found that the FSH pathway was activated by ING5, with ING5 overexpression increasing levels of membrane bound FSH receptors. Blocking the FSH pathway also reduced the ability of BTICs to self-renew. In ovarian cancer cells FSH activates OCT4 expression through the ERK pathway.69 Here we found that FSH treatment increased OCT4 expression in BTICs, suggesting a mechanism shared in both cell types. ING5 also activated a network of genes involved in hormone signaling and steroidogenesis (Supplementary Figure S3d). Among these, NR5A1 (Figure 6c) has a central function in regulating steroidogenesis and the expression of a variety of hormones. NR5A1 also helps induce a ground state of pluripotency and OCT4 expression82, 83 suggesting that ING5 has a function in activating the steroidogenesis-hormone network universally in pluripotent and cancer stem cell systems.
ING5 has more often been described as a tumor suppressor although some multi-faceted effects were also observed, such as inhibition of chemoresistance and apoptosis.37, 43 In contrast, our study shows that ING5 plays an oncogenic role in brain tumors by enhancing the self-renewal of stem-like cancer cells. It is clear from previous studies that the functions of ING5 are highly dependent on its interaction partners. In this study since the major cell line used (BT 189) harbors homozygous p53 mutation R273C which greatly alters its target gene pattern,73, 84 we predict that the role of ING5 is not dependent on p53, which is consistent with an undisturbed cell cycle profile in ING5-manipulated cells.
Important roles of HAT enzymes and cofactors in the ING5–HAT complexes have been documented in mammalian brain development. Disruption of acetyltransferase MORF causes severe defects in brain development and adult neurogenesis in mice,52, 85 and is associated with multiple neurodevelopmental disorders in human.86, 87, 88 Deficiency of BRPF1, a cofactor present in the same complexes as ING5, leads to neocortical and hippocampus abnormalities of mice.89, 90 These complexes are believed to play a critical role in promoting neural stem cell characteristics,91, 92 however an understanding of the mechanism(s) by which they promote stem cell character and correlation with brain tumor development is still lacking. Our results highlight the requirement of ING5 for maintaining BTIC stem cell properties, suggesting that proper targeting of associated complexes is critical for maintaining stemness.
In summary, we find that ING5 promotes stem cell properties of BTICs by inducing calcium and FSH signaling pathways, which stimulate PI3K/AKT and MEK/ERK signaling. The stem cell factors OCT4, OLIG2 and Nestin were subsequently activated. Our data suggest FSH and calcium signaling primarily function through distinct mechanisms, involving the CaN axis and the activation of OCT4, respectively. These effects were dependent on the PHD motif of ING5, which targets HAT complexes to activate gene expression as outlined in the model shown in Figure 7h. BTICs are the drivers of tumor initiation and recurrence in GBMs and are difficult to eradicate due to their stem cell properties. We suggest that besides pharmaceutically targeting ING5 itself, the FSH and calcium signaling pathway could also serve as targets and may prove useful in combination with the current PI3K and MEK pathway inhibitors being tested, to diminish stem cell properties, and hence the propensity for BTIC-based GBM to recur.
Materials and methods
BTIC cultures and sphere formation assays
BTIC lines BT 12, BT 134 and BT 189 were established from adult GBM patients as described previously.6 Neurospheres were kept in serum free media and supplemented with 20 ng/ml EGF (Peprotech, Rocky Hill, NJ, USA), 20 ng/ml FGF (R&D Systems, Minneapolis, MN, USA) and 2 μg/ml heparin solution (STEMCELL, Vancouver BC, CA, USA). For sphere formation assays, 200 viable single cells were grown for 12–14 days in each well of 96-well plates. The complete field of each well was captured under a × 4 microscope objective and the diameter of each sphere measured. At least three wells (600 cells) were counted for each condition in one experiment. Serial sphere assays were conducted by harvesting all of the cells from the last passage and replating them into new wells at the same dilution. Assays with treatment by protein kinase inhibitors, calcium modulators and other reagents were conducted in the absence of growth factors. All statistical analyses were done by Student t-test (unpaired, two-sided).
Sphere cell differentiation and immunofluorescence
BTICs were plated on poly-l-ornithine coated glass coverslips and induced to differentiate in serum free media supplemented with 1% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) for 5–7 days. For immunofluorescence assays, cells were fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100 and then incubated with the primary antibodies against Nestin (1: 800; MAB-1259, R&D Systems), Tubb3 (1:400; MRB-435P, Biolegend, San Diego, CA, USA), GFAP (1:200; Z0334, Dako, Glostrup, Denmark) or FSHR (1:30, MAB65591, R&D Systems) at 4 °C overnight as described.93 After incubation with fluorophore-conjugated secondary antibodies (Invitrogen) and Hoechst 33258 (Sigma, St Louis, MO, USA) at 37 °C, coverslips were examined under an Olympus IX71 wide-field microscope. At least three independent experiments were performed and representative fields are shown in the immunofluorescence results.
Asymmetry studies
The division mode analysis was performed on mitotic pairs seeded at low density on coated coverslips. Cells were synchronized using the double thymidine block method and released for 18–20 h before being fixed and immunostaining for Nestin.
Plasmid transfection and PiggyBac overexpression
The pCI-ING5 plasmid was constructed by cloning ING5 cDNA into the pCI empty vector (Promega, Madison, WI, USA). pCI-ING5 and pCI vector were transfected into BTICs using lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. To establish stable overexpression cell lines, the PiggyBac vector (PBQM530A, Systems Biosciences, Palo Alto, CA, USA) containing ING5 cDNA was co-transfected with the transposase plasmid (PB210PA-1, Systems Biosciences) into BT 189 cells. Positive clones were selected with puromycin. Expression of ING5 was induced by cumate (Sigma) at 30 μg/ml (1 ×). The PHD motif (186–240 aa) was removed from the ING5 sequence in the iPB-ING5-ΔPHD construction. A FLAG tag (DYKDDDDK) was added to the c-terminus of full-length or truncated ING5 proteins in iPB-ING5-FLAG and iPB-ING5-ΔPHD constructions.
siRNA and lentiviral-based shRNA systems
This information is provided in the Expanded View section.
Quantitative real-time PCR
Total RNA was isolated using TRIzol (Invitrogen) reagent and reverse transcription was conducted using the Applied Biosystems (Foster City, CA, USA) cDNA Reverse Transcription kit. Real-time qPCR reactions were carried out using Maxima SYBR Green qPCR Mastermix (Fermentas, Waltham, MA, USA) on the QuantStudio 6 Flex real-time PCR system (Applied Biosystems).
Gene expression microarray and data analysis
This information is provided in the Expanded View section. Primary data are available through the GEO access number GSE100398 that can be viewed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100398.
Antibodies and chemicals
This information is provided in the Expanded View section.
Flow cytometry
Live cells were incubated with conjugated antibodies CD133-PE (1:100; 12-1338, eBioscience, San Diego, CA, USA) and CD44-FITC (1:50; ab19622, abcam, Cambridge, UK) at 4 °C for 30 min. PE Mouse IgG1 kapa (400111, Biolegend) and FITC Rat IgG2b (400633, Biolegend) were used as isotype controls. After washing with PBS, cells were re-suspended in 300 μl PBS and examined on an LSR II flow cytometer (BD Biosciences, San Jose, CA, USA). Data analysis was performed using FlowJo software (Tree Star, Inc., Ashland, OR, USA). For calcium detection, live cells were loaded with 5 μM Fluo3-AM (Sigma) for 45 min at 37 °C, washed with PBS, and checked on the flow cytometer. For cell cycle analysis, transfected cells were dissociated into single cell suspension and fixed in 70% ethanol. After washing with PBS, cells were treated with RNase (0.2 mg/ml) and stained in propidium iodide solution (10 μg/ml) and then analyzed by flow cytometry.
Live cell calcium imaging
Cells were cultured on poly-L-ornithine coated dishes overnight in the absence of growth factors, and then incubated in medium containing 5 μM Fluo3-AM at 37 °C for 30 min. After washing with PBS, cells were de-esterified for 30 min and examined under a fluorescence microscope. For co-transfection experiments (Supplementary Figure S4a), the calcium level was quantified using ImageJ software.
Chromatin immunoprecipitation
About 5 × 106 BTICs were used for each IP condition which were first dissociated and crosslinked with 1% formaldehyde. Cells were lysed in ChIP lysis buffer as described previously.94 Extracts were sonicated for 10 times (10 s each). An aliquot of 50 μl of lysates were kept as input controls and the rest were incubated overnight with the ING5 antibody, FLAG M2 Affinity Gel (A2220, Sigma), rabbit/mouse IgG controls (Santa Cruz, Dallas, TX, USA), respectively, followed by protein A and protein G Sepharose (GE Healthcare, Little Chalfont, UK) incubation for 2 h (except for the M2-gel treated samples). Agarose Beads were then washed and eluted as described.94 The eluate was de-crosslinked and DNA isolated using phenol-chloroform and ethanol.
Statistics
All t-tests were performed in GraphPad Prism and results were displayed as the mean±s.e.m. At least three independent experiments were performed to obtain adequate power for the statistical analyses. Linear regression, Pearson Correlation and Kaplan Meier analyses were performed using SPSS Statistics software (IBM, Armonk, NY, USA). For the overall Kaplan Meier analysis on TCGA data, all available GBM cases (n=114) were included and for subtype-specific analyses, G-CIMP subtype was excluded due to small sample size (n=6). The gene expression data from TCGA GBM cohorts were not assumed to be normally distributed and the Pearson and linear regression analyses were performed accordingly.
GEO access. Primary data are available through the GEO access number GSE100398 that can be viewed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100398.
References
Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol 2015; 17: iv1–iv62.
Omuro A, DeAngelis LM . Glioblastoma and other malignant gliomas: a clinical review. JAMA 2013; 310: 1842–1850.
Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA . Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002; 39: 193–206.
Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003; 63: 5821–5828.
Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–7021.
Kelly JJ, Stechishin O, Chojnacki A, Lun X, Sun B, Senger DL et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells 2009; 27: 1722–1733.
Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012; 488: 522–526.
Gao X, McDonald JT, Hlatky L, Enderling H . Acute and fractionated irradiation differentially modulate glioma stem cell division kinetics. Cancer Res 2013; 73: 1481–1490.
Swartling FJ, Cancer M, Frantz A, Weishaupt H, Persson AI . Deregulated proliferation and differentiation in brain tumors. Cell Tissue Res 2015; 359: 225–254.
Niola F, Zhao X, Singh D, Sullivan R, Castano A, Verrico A et al. Mesenchymal high-grade glioma is maintained by the ID-RAP1 axis. J Clin Investig 2013; 123: 405–417.
Zhao N, Guo Y, Zhang M, Lin L, Zheng Z . Akt-mTOR signaling is involved in Notch-1-mediated glioma cell survival and proliferation. Oncol Rep 2010; 23: 1443–1447.
Ross SE, Greenberg ME, Stiles CD . Basic helix-loop-helix factors in cortical development. Neuron 2003; 39: 13–25.
Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 2002; 1: 269–277.
Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC . PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev 2001; 15: 1913–1925.
Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006; 9: 391–403.
Fullgrabe J, Kavanagh E, Joseph B . Histone onco-modifications. Oncogene 2011; 30: 3391–3403.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–477.
Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K . Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs. Genes Dis 2015; 2: 152–163.
Caren H, Stricker SH, Bulstrode H, Gagrica S, Johnstone E, Bartlett TE et al. Glioblastoma stem cells respond to differentiation cues but fail to undergo commitment and terminal cell-cycle arrest. Stem Cell Rep 2015; 5: 829–842.
Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014; 157: 580–594.
Signaroldi E, Laise P, Cristofanon S, Brancaccio A, Reisoli E, Atashpaz S et al. Polycomb dysregulation in gliomagenesis targets a Zfp423-dependent differentiation network. Nat Commun 2016; 7: 10753.
Heddleston JM, Wu Q, Rivera M, Minhas S, Lathia JD, Sloan AE et al. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ 2012; 19: 428–439.
Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV . The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 1998; 391: 295–298.
Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP et al. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci 2001; 114: 3455–3462.
Schafer A, Karaulanov E, Stapf U, Doderlein G, Niehrs C . Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes Dev 2013; 27: 261–273.
Garkavtsev I, Kazarov A, Gudkov A, Riabowol K . Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet 1996; 14: 415–420.
Vieyra D, Loewith R, Scott M, Bonnefin P, Boisvert FM, Cheema P et al. Human ING1 proteins differentially regulate histone acetylation. J Biol Chem 2002; 277: 29832–29839.
Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 2006; 21: 51–64.
Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D . Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol 2000; 20: 3807–3816.
Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006; 442: 100–103.
Tallen G, Riabowol K . Keep-ING balance: tumor suppression by epigenetic regulation. FEBS Lett 2014; 588: 2728–2742.
Saito M, Kumamoto K, Robles AI, Horikawa I, Furusato B, Okamura S et al. Targeted disruption of Ing2 results in defective spermatogenesis and development of soft-tissue sarcomas. PLoS ONE 2010; 5: e15541.
Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T, Imai H, Minami N . ING3 is essential for asymmetric cell division during mouse oocyte maturation. PLoS ONE 2013; 8: e74749.
Eapen SA, Netherton SJ, Sarker KP, Deng L, Chan A, Riabowol K et al. Identification of a novel function for the chromatin remodeling protein ING2 in muscle differentiation. PLoS ONE 2012; 7: e40684.
Linzen U, Lilischkis R, Pandithage R, Schilling B, Ullius A, Luscher-Firzlaff J et al. ING5 is phosphorylated by CDK2 and controls cell proliferation independently of p53. PLoS ONE 2015; 10: e0123736.
Zhang F, Baumer N, Rode M, Ji P, Zhang T, Berdel WE et al. The inhibitor of growth protein 5 (ING5) depends on INCA1 as a co-factor for its antiproliferative effects. PLoS ONE 2011; 6: e21505.
Gou WF, Shen DF, Yang XF, Zhao S, Liu YP, Sun HZ et al. ING5 suppresses proliferation, apoptosis, migration and invasion, and induces autophagy and differentiation of gastric cancer cells: a good marker for carcinogenesis and subsequent progression. Oncotarget 2015; 6: 19552–19579.
Zhang F, Zhang X, Meng J, Zhao Y, Liu X, Liu Y et al. ING5 inhibits cancer aggressiveness via preventing EMT and is a potential prognostic biomarker for lung cancer. Oncotarget 2015; 6: 16239–16252.
Mendes-Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci USA 2012; 109: 2730–2735.
Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res 2003; 63: 2373–2378.
Liu N, Wang J, Wang R, Liu Z, Yu Y, Lu H . ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res 2013; 73: 3749–3760.
Ding XQ, Zhao S, Yang L, Zhao X, Zhao GF, Zhao SP et al. The nucleocytoplasmic translocation and up-regulation of ING5 protein in breast cancer: a potential target for gene therapy. Oncotarget 2017; doi: 10.18632/oncotarget.17918.
Zhao S, Zhao ZJ, He HY, Wu JC, Ding XQ, Yang L et al. The roles of ING5 in gliomas: a good marker for tumorigenesis and a potential target for gene therapy. Oncotarget 2017; 8: 56558–56568.
Zhao S, Yang XF, Shen DF, Gao Y, Shi S, Wu JC et al. The down-regulated ING5 expression in lung cancer: a potential target of gene therapy. Oncotarget 2016; 7: 54596–54615.
Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C et al. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol 2008; 28: 6828–6843.
Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol 2012; 14: 753–763.
Vlismas A, Bletsa R, Mavrogianni D, Mamali G, Pergamali M, Dinopoulou V et al. Microarray analyses reveal marked differences in growth factor and receptor expression between 8-cell human embryos and pluripotent stem cells. Stem Cells Dev 2016; 25: 160–177.
Hussein SM, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL et al. Genome-wide characterization of the routes to pluripotency. Nature 2014; 516: 198–206.
Sarkar P, Mischler A, Randall SM, Collier TS, Dorman KF, Boggess KA et al. Identification of epigenetic factor proteins expressed in human embryonic stem cell-derived trophoblasts and in human placental trophoblasts. J Proteome Res 2016; 15: 2433–2444.
Kim MS, Cho HI, Park SH, Kim JH, Chai YG, Jang YK . The histone acetyltransferase Myst2 regulates Nanog expression, and is involved in maintaining pluripotency and self-renewal of embryonic stem cells. FEBS Lett 2015; 589: 941–950.
Perez-Campo FM, Borrow J, Kouskoff V, Lacaud G . The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood 2009; 113: 4866–4874.
Merson TD, Dixon MP, Collin C, Rietze RL, Bartlett PF, Thomas T et al. The transcriptional coactivator Querkopf controls adult neurogenesis. J Neurosci 2006; 26: 11359–11370.
Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014; 14: 357–369.
Liu Q, Nguyen DH, Dong Q, Shitaku P, Chung K, Liu OY et al. Molecular properties of CD133+ glioblastoma stem cells derived from treatment-refractory recurrent brain tumors. J Neurooncol 2009; 94: 1–19.
Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009; 458: 766–770.
Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol 2008; 10: 93–101.
Kosodo Y, Roper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB . Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J 2004; 23: 2314–2324.
Chang KC, Wang C, Wang H . Balancing self-renewal and differentiation by asymmetric division: insights from brain tumor suppressors in Drosophila neural stem cells. BioEssays 2012; 34: 301–310.
Ayuso-Sacido A, Moliterno JA, Kratovac S, Kapoor GS, O'Rourke DM, Holland EC et al. Activated EGFR signaling increases proliferation, survival, and migration and blocks neuronal differentiation in post-natal neural stem cells. J Neurooncol 2010; 97: 323–337.
Reynolds BA, Weiss S . Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 1992; 255: 1707–1710.
Lawn S, Krishna N, Pisklakova A, Qu X, Fenstermacher DA, Fournier M et al. Neurotrophin signaling via TrkB and TrkC receptors promotes the growth of brain tumor-initiating cells. J Biol Chem 2015; 290: 3814–3824.
Luberg K, Park R, Aleksejeva E, Timmusk T . Novel transcripts reveal a complex structure of the human TRKA gene and imply the presence of multiple protein isoforms. BMC Neurosci 2015; 16: 78.
Ishiuchi S, Tsuzuki K, Yoshida Y, Yamada N, Hagimura N, Okado H et al. Blockage of Ca(2+)-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat Med 2002; 8: 971–978.
Kang SS, Han KS, Ku BM, Lee YK, Hong J, Shin HY et al. Caffeine-mediated inhibition of calcium release channel inositol 1,4,5-trisphosphate receptor subtype 3 blocks glioblastoma invasion and extends survival. Cancer Res 2010; 70: 1173–1183.
Chai S, Xu X, Wang Y, Zhou Y, Zhang C, Yang Y et al. Ca2+/calmodulin-dependent protein kinase IIgamma enhances stem-like traits and tumorigenicity of lung cancer cells. Oncotarget 2015; 6: 16069–16083.
Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM et al. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 2011; 472: 366–369.
Bhartiya D, Singh J . FSH-FSHR3-stem cells in ovary surface epithelium: basis for adult ovarian biology, failure, aging, and cancer. Reproduction 2015; 149: R35–R48.
Zhang Z, Zhu Y, Lai Y, Wu X, Feng Z, Yu Y et al. Follicle-stimulating hormone inhibits apoptosis in ovarian cancer cells by regulating the OCT4 stem cell signaling pathway. Int J Oncol 2013; 43: 1194–1204.
Liu L, Zhang J, Fang C, Zhang Z, Feng Y, Xi X . OCT4 mediates FSH-induced epithelial-mesenchymal transition and invasion through the ERK1/2 signaling pathway in epithelial ovarian cancer. Biochem Biophys Res Commun 2015; 461: 525–532.
Siegel ET, Kim HG, Nishimoto HK, Layman LC . The molecular basis of impaired follicle-stimulating hormone action: evidence from human mutations and mouse models. Reprod Sci 2013; 20: 211–233.
Flores JA, Veldhuis JD, Leong DA . Follicle-stimulating hormone evokes an increase in intracellular free calcium ion concentrations in single ovarian (granulosa) cells. Endocrinology 1990; 127: 3172–3179.
Kataoka H, Bonnefin P, Vieyra D, Feng X, Hara Y, Miura Y et al. ING1 represses transcription by direct DNA binding and through effects on p53. Cancer Res 2003; 63: 5785–5792.
Cusulin C, Chesnelong C, Bose P, Bilenky M, Kopciuk K, Chan JA et al. Precursor states of brain tumor initiating cell lines are predictive of survival in xenografts and associated with glioblastoma subtypes. Stem Cell Rep 2015; 5: 1–9.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014; 344: 1396–1401.
Wegner M, Stolt CC . From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci 2005; 28: 583–588.
Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN . Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 2000; 25: 55–57.
Wang Y, Kim E, Wang X, Novitch BG, Yoshikawa K, Chang LS et al. ERK inhibition rescues defects in fate specification of Nf1-deficient neural progenitors and brain abnormalities. Cell 2012; 150: 816–830.
Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell 2008; 13: 69–80.
Davis FB, Tang HY, Shih A, Keating T, Lansing L, Hercbergs A et al. Acting via a cell surface receptor, thyroid hormone is a growth factor for glioma cells. Cancer Res 2006; 66: 7270–7275.
Velasco I . Stem cells with neurogenic potential and steroid hormones. Curr Top Med Chem 2011; 11: 1684–1693.
Liu L, Wang J, Zhao L, Nilsen J, McClure K, Wong K et al. Progesterone increases rat neural progenitor cell cycle gene expression and proliferation via extracellularly regulated kinase and progesterone receptor membrane components 1 and 2. Endocrinology 2009; 150: 3186–3196.
Guo G, Smith A . A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 2010; 137: 3185–3192.
Yang HM, Do HJ, Kim DK, Park JK, Chang WK, Chung HM et al. Transcriptional regulation of human Oct4 by steroidogenic factor-1. J Cell Biochem 2007; 101: 1198–1209.
Peng Y, Li C, Chen L, Sebti S, Chen J . Rescue of mutant p53 transcription function by ellipticine. Oncogene 2003; 22: 4478–4487.
Thomas T, Voss AK, Chowdhury K, Gruss P . Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 2000; 127: 2537–2548.
Kraft M, Cirstea IC, Voss AK, Thomas T, Goehring I, Sheikh BN et al. Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice. J Clin Investig 2011; 121: 3479–3491.
Clayton-Smith J, O'Sullivan J, Daly S, Bhaskar S, Day R, Anderson B et al. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am J Hum Genet 2011; 89: 675–681.
Simpson MA, Deshpande C, Dafou D, Vissers LE, Woollard WJ, Holder SE et al. De novo mutations of the gene encoding the histone acetyltransferase KAT6B cause Genitopatellar syndrome. Am J Hum Genet 2012; 90: 290–294.
You L, Zou J, Zhao H, Bertos NR, Park M, Wang E et al. Deficiency of the chromatin regulator BRPF1 causes abnormal brain development. J Biol Chem 2015; 290: 7114–7129.
You L, Yan K, Zou J, Zhao H, Bertos NR, Park M et al. The lysine acetyltransferase activator Brpf1 governs dentate gyrus development through neural stem cells and progenitors. PLoS Genet 2015; 11: e1005034.
Sheikh BN, Dixon MP, Thomas T, Voss AK . Querkopf is a key marker of self-renewal and multipotency of adult neural stem cells. J Cell Sci 2012; 125: 295–309.
Perez-Campo FM, Costa G, Lie ALM, Stifani S, Kouskoff V, Lacaud G . MOZ-mediated repression of p16(INK) (4) (a) is critical for the self-renewal of neural and hematopoietic stem cells. Stem Cells 2014; 32: 1591–1601.
Vieyra D, Senger DL, Toyama T, Muzik H, Brasher PM, Johnston RN et al. Altered subcellular localization and low frequency of mutations of ING1 in human brain tumors. Clin Cancer Res 2003; 9: 5952–5961.
Rajarajacholan UK, Thalappilly S, Riabowol K . The ING1a tumor suppressor regulates endocytosis to induce cellular senescence via the Rb-E2F pathway. PLoS Biol 2013; 11: e1001502.
Acknowledgements
We are grateful to Dr. Samuel Weiss’s group, particularly Dr. Artee Luchman and Rozina Hassam, from the University of Calgary for providing the patient-derived BTIC lines and technical support. We thank Dr. Jennifer Cobb and Dr. Derrick Rancourt from the University of Calgary for valuable experimental insights. This work was funded by the Alberta Cancer Foundation (Project 26512) and Canadian Institutes of Health Research (MOP-133646). This work was funded by grants from the Alberta Cancer Foundation (Project 26512) and Canadian Institutes of Health Research (MOP-133646) to KR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Wang, F., Wang, A., Chesnelong, C. et al. ING5 activity in self-renewal of glioblastoma stem cells via calcium and follicle stimulating hormone pathways. Oncogene 37, 286–301 (2018). https://doi.org/10.1038/onc.2017.324
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.324
This article is cited by
-
Molecular mechanisms of inhibitor of growth (ING) family members in health and malignancy
Cancer Cell International (2022)
-
Inhibitor of Growth 4 (ING4) is a positive regulator of rRNA synthesis
Scientific Reports (2019)
-
Oligonucleotide Therapeutics as a New Class of Drugs for Malignant Brain Tumors: Targeting mRNAs, Regulatory RNAs, Mutations, Combinations, and Beyond
Neurotherapeutics (2019)