Abstract
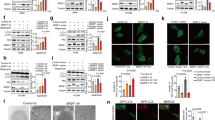
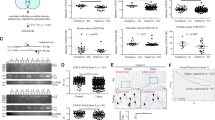
Resisting death is a central hallmark of cancer cells. Tumors rely on a number of genetic mechanisms to avoid apoptosis, and alterations in mRNA alternative splicing are increasingly recognized to have a role in tumorigenesis. In this study, we identify the splicing regulator SLU7 as an essential factor for the preservation of hepatocellular carcinoma (HCC) cells viability. Compared with hepatocytes, SLU7 expression is reduced in HCC cells; however, further SLU7 depletion triggered autophagy-related cellular apoptosis in association with the overproduction of reactive oxygen species. Remarkably, these responses were not observed in primary human hepatocytes or in the well-differentiated HepaRG cell line. Mechanistically, we demonstrate that SLU7 binds the C13orf25 primary transcript in which the polycistronic oncomir miR-17-92 cluster is encompassed, and is necessary for its processing and expression. SLU7 knockdown altered the splicing of the C13orf25 primary transcript, and markedly reduced the expression of its miR-17, miR-20 and miR-92a constituents. This led to the upregulation of CDKN1A (P21) and BCL2L11 (BIM) expression, two bona fide targets of the miR-17-92 cluster and recognized mediators of its pro-survival and tumorigenic activity. Interestingly, altered splicing of miR-17-92 and downregulation of miR-17 and miR-20 were not observed upon SLU7 knockdown in non-transformed hepatocytes, but was found in other (HeLa, H358) but not in all (Caco2) non-hepatic tumor cells. The functional relevance of miR-17-92 dysregulation upon SLU7 knockdown was established when oxidative stress, autophagy and apoptosis were reversed by co-transfection of HCC cells with a miR-17 mimic. Together, these findings indicate that SLU7 is co-opted by HCC cells and other tumor cell types to maintain survival, and identify this splicing regulator as a new determinant for the expression of the oncogenic miR-17-92 cluster. This novel mechanism may be exploited for the development of antitumoral strategies in cancers displaying such SLU7-miR-17-92 crosstalk.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lee Y, Rio DC . Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 2015; 84: 291–323.
David CJ, Manley JL . Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 2010; 24: 2343–2364.
Singh RK, Cooper TA . Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 2012; 18: 472–482.
Faustino NA, Cooper TA . Pre-mRNA splicing and human disease. Genes Dev 2003; 17: 419–437.
Castillo J, Goñi S, Latasa MU, Perugorría MJ, Calvo A, Muntané J et al. Amphiregulin induces the alternative splicing of p73 into its oncogenic isoform DeltaEx2p73 in human hepatocellular tumors. Gastroenterology 2009; 137: 1805–1815.
Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Clapéron A, Chrétien Y et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res 2013; 73: 3974–3986.
Berasain C, Elizalde M, Urtasun R, Castillo J . Alterations in the expression and activity of pre-mRNA splicing factors in hepatocarcinogenesis. Hepatic Oncology 2014; 1: 241–252.
David CJ, Chen M, Assanah M, Canoll P, Manley JL . HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010; 463: 364–368.
Elizalde M, Urtasun R, Azkona M, Latasa MU, Goñi S, Garcia-Irigoyen O et al. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest 2014; 124: 2909–2920.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S et al. A microRNA polycistron as a potential human oncogene. Nature 2005; 435: 828–833.
Mendell JT . miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008; 133: 217–222.
Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N . Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta 2013; 1833: 3448–3459.
Jiang X, Overholtzer M, Thompson CB . Autophagy in cellular metabolism and cancer. J Clin Invest 2015; 125: 47–54.
Filomeni G, De Zio D, Cecconi F . Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ 2015; 22: 377–388.
Ebi H, Sato T, Sugito N, Hosono Y, Yatabe Y, Matsuyama Y et al. Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene 2009; 28: 3371–3379.
Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 2004; 64: 3087–3095.
Olive V, Jiang I, He L . mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010; 42: 1348–1354.
Thomas M, Lange-Grünweller K, Hartmann D, Golde L, Schlereth J, Streng D et al. Analysis of Transcriptional Regulation of the Human miR-17-92 Cluster; Evidence for Involvement of Pim-1. Int J Mol Sci 2013; 14: 12273–12296.
Concepcion CP, Bonetti C, Ventura A . The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J 2012; 18: 262–267.
Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008; 13: 272–286.
Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE 2008; 3: e2236.
Inomata M, Tagawa H, Guo Y-M, Kameoka Y, Takahashi N, Sawada K . MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood 2009; 113: 396–402.
Olive V, Li Q, He L . mir-17-92: a polycistronic oncomir with pleiotropic functions. Immunol Rev 2013; 253: 158–166.
Mogilyansky E, Rigoutsos I . The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ 2013; 20: 1603–1614.
Li Y, Choi PS, Casey SC, Dill DL, Felsher DW . MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 2014; 26: 262–272.
Osada H, Takahashi T . let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci 2011; 102: 9–17.
Alberstein M, Amit M, Vaknin K, O'Donnell A, Farhy C, Lerenthal Y et al. Regulation of transcription of the RNA splicing factor hSlu7 by Elk-1 and Sp1 affects alternative splicing. RNA 2007; 13: 1988–1999.
Chen J, Weiss WA . Alternative splicing in cancer: implications for biology and therapy. Oncogene 2015; 34: 1–14.
Oltean S, Bates DO . Hallmarks of alternative splicing in cancer. Oncogene 2014; 33: 5311–5318.
Koh CM, Bezzi M, Low DHP, Ang WX, Teo SX, Gay FPH et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015; 523: 96–100.
Bellot GL, Liu D, Pervaiz S . ROS, autophagy, mitochondria and cancer: Ras, the hidden master? Mitochondrion 2013; 13: 155–162.
Czaja MJ, Ding W-X, Donohue TM, Friedman SL, Kim J-S, Komatsu M et al. Functions of autophagy in normal and diseased liver. autophagy 2013; 9: 1131–1158.
Das S, Krainer AR . Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 2014; 12: 1195–1204.
Ghigna C, Moroni M, Porta C, Riva S, Biamonti G . Altered expression of heterogeneous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res 1998; 58: 5818–5824.
Kurokawa K, Akaike Y, Masuda K, Kuwano Y, Nishida K, Yamagishi N et al. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene 2014; 33: 1407–1417.
Tan W, Li Y, Lim SG, Tan TMC . miR-106b-25/miR-17-92 clusters: polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol 2014; 20: 5962–5972.
Zhu H, Han C, Lu D, Wu T . miR-17-92 cluster promotes cholangiocarcinoma growth: evidence for PTEN as downstream target and IL-6/Stat3 as upstream activator. Am J Pathol 2014; 184: 2828–2839.
Yang H, Lan P, Hou Z, Guan Y, Zhang J, Xu W et al. Histone deacetylase inhibitor SAHA epigenetically regulates miR-17-92 cluster and MCM7 to upregulate MICA expression in hepatoma. Br J Cancer 2015; 112: 112–121.
Ji M, Rao E, Ramachandrareddy H, Shen Y, Jiang C, Chen J et al. The miR-17-92 microRNA cluster is regulated by multiple mechanisms in B-cell malignancies. Am J Pathol 2011; 179: 1645–1656.
Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005; 65: 9628–9632.
Shomron N, Levy C . MicroRNA-biogenesis and Pre-mRNA splicing crosstalk. J Biomed Biotechnol 2009; 2009: 594678.
Janas MM, Khaled M, Schubert S, Bernstein JG, Golan D, Veguilla RA et al. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet 2011; 7: e1002330.
Ying SY, Lin SL . Current perspectives in intronic micro RNAs (miRNAs). J Biomed Sci 2006; 13: 5–15.
Guil S, Caceres JF . The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 2007; 14: 591–596.
Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR et al. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res 2009; 69: 8844–8852.
Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther 2013; 14: 574–586.
Macip S, Igarashi M, Fang L, Chen A, Pan ZQ, Lee SW et al. Inhibition of p21-mediated ROS accumulation can rescue p21-induced senescence. EMBO J 2002; 21: 2180–2188.
Hsu TYT, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015; 525: 384–388.
Pichard L, Raulet E, Fabre G, Ferrini JB, Ourlin J-C, Maurel P . Human hepatocyte culture. Methods Mol Biol 2006; 320: 283–293.
Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res 2001; 61: 439–444.
Castillo J, Erroba E, Perugorría MJ, Santamaría M, Lee DC, Prieto J et al. Amphiregulin contributes to the transformed phenotype of human hepatocellular carcinoma cells. Cancer Res 2006; 66: 6129–6138.
Berasain C, García-Trevijano ER, Castillo J, Erroba E, Santamaría M, Lee DC et al. Novel role for amphiregulin in protection from liver injury. J Biol Chem 2005; 280: 19012–19020.
Boya P, González-Polo R-A, Casares N, Perfettini J-L, Dessen P, Larochette N et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005; 25: 1025–1040.
Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S . Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 2004; 304: 1500–1502.
Perugorría MJ, Castillo J, Latasa MU, Goñi S, Segura V, Sangro B et al. Wilms' tumor 1 gene expression in hepatocellular carcinoma promotes cell dedifferentiation and resistance to chemotherapy. Cancer Res 2009; 69: 1358–1367.
Urtasun R, Latasa MU, Demartis MI, Balzani S, Goñi S, Garcia-Irigoyen O et al. Connective tissue growth factor autocriny in human hepatocellular carcinoma: oncogenic role and regulation by epidermal growth factor receptor/yes-associated protein-mediated activation. Hepatology 2011; 54: 2149–2158.
Acknowledgements
FIMA and the ‘UTE project CIMA’; RTICC-RD06 00200061; CIBERehd; Grant numbers: FIS PI10/02642, PI13/00359, PI10/00038, PI13/00385 from Instituto de Salud Carlos III (ISCIII) co-financed by ‘Fondo Europeo de Desarrollo Regional’ (FEDER) ‘Una manera de hacer Europa’; Gobierno de Navarra; ‘Torres Quevedo’ and ‘Ramón y Cajal’ contracts from Ministerio de Educación; PFIS fellowship from ISCIII; FPU fellowship from Ministerio de Educación, Cultura y Deporte; Fundaciónes Mario Losantos, MTorres, Eugenio Rodríguez Pascual; Agustina Damboriena Support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Urtasun, R., Elizalde, M., Azkona, M. et al. Splicing regulator SLU7 preserves survival of hepatocellular carcinoma cells and other solid tumors via oncogenic miR-17-92 cluster expression. Oncogene 35, 4719–4729 (2016). https://doi.org/10.1038/onc.2015.517
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.517
This article is cited by
-
Splicing factor SRSF3 represses translation of p21cip1/waf1 mRNA
Cell Death & Disease (2022)
-
SLC38A4 functions as a tumour suppressor in hepatocellular carcinoma through modulating Wnt/β-catenin/MYC/HMGCS2 axis
British Journal of Cancer (2021)
-
MicroRNA-17-92 cluster promotes the proliferation and the chemokine production of keratinocytes: implication for the pathogenesis of psoriasis
Cell Death & Disease (2018)
-
miR-363-5p as potential prognostic marker for hepatocellular carcinoma indicated by weighted co-expression network analysis of miRNAs and mRNA
BMC Gastroenterology (2017)