Abstract
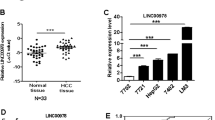
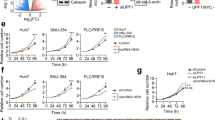
Long non-coding RNAs (lncRNA) play crucial roles in hepatocellular carcinoma (HCC) progression. However, the specific functions of lncRNAs in alternative splicing (AS) and the metastatic cascade in liver cancer remain largely unclear. In this study, we identified a novel lncRNA, LINC01348, which was significantly downregulated in HCC and correlated with survival functions in HCC patients. Ectopic expression of LINC01348 induced marked inhibition of cell growth, and metastasis in vitro and in vivo. Conversely, these phenotypes were reversed upon knockdown of LINC01348. Mechanistically, LINC01348 complexed with splicing factor 3b subunit 3 (SF3B3) acted as a modulator of EZH2 pre-mRNA AS, and induced alterations in JNK/c-Jun activity and expression of Snail. Notably, C-terminal truncated HBx (Ct-HBx) negatively regulated LINC01348 through c-Jun signaling. Our data collectively highlight those novel regulatory associations involving LINC01348/SF3B3/EZH2/JNK/c-Jun/Snail are an important determinant of metastasis in HCC cells and support the potential utility of targeting LINC01348 as a therapeutic strategy for HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lim JK, Nguyen MH, Kim WR, Gish R, Perumalswami P, Jacobson IM. Prevalence of Chronic Hepatitis B Virus Infection in the United States. Am J Gastroenterol. 2020;115:1429–38.
Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250–61.
Yeh CT, Lai MW. Eliminating hepatitis B virus through neonatal vaccination: can we make it? J Hepatol. 2012;57:484–5.
Shi J, Zhu L, Liu S, Xie WF. A meta-analysis of case-control studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma in China. Br J Cancer. 2005;92:607–12.
Donato F, Boffetta P, Puoti M. A meta-analysis of epidemiological studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma. Int J Cancer. 1998;75:347–54.
Sze KM, Chu GK, Lee JM, Ng IO. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology. 2013;57:131–9.
Quetier I, Brezillon N, Revaud J, Ahodantin J, DaSilva L, Soussan P, et al. C-terminal-truncated hepatitis B virus X protein enhances the development of diethylnitrosamine-induced hepatocellular carcinogenesis. J Gen Virol. 2015;96:614–25.
Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, Pavarino EC, da Silva RF, da Silva RC, et al. Hepatocellular carcinoma: a comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac J Cancer Prev. 2017;18:863–72.
Lurje I, Czigany Z, Bednarsch J, Roderburg C, Isfort P, Neumann UP. et al. Treatment Strategies for Hepatocellular Carcinoma (-) a Multidisciplinary Approach. Int J Mol Sci. 2019;20:1465.
Park JW, Kim YJ, Kim DY, Bae SH, Paik SW, Lee YJ, et al. Sorafenib with or without concurrent transarterial chemoembolization in patients with advanced hepatocellular carcinoma: the phase III STAH trial. J Hepatol. 2019;70:684–91.
Uchino K, Tateishi R, Shiina S, Kanda M, Masuzaki R, Kondo Y, et al. Hepatocellular carcinoma with extrahepatic metastasis: clinical features and prognostic factors. Cancer. 2011;117:4475–83.
Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–66.
Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110.
Ghafouri-Fard S, Taheri M. Nuclear Enriched Abundant Transcript 1 (NEAT1): a long non-coding RNA with diverse functions in tumorigenesis. Biomed Pharmacother. 2019;111:51–9.
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38.
Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, et al. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380:476–84.
Hodo Y, Honda M, Tanaka A, Nomura Y, Arai K, Yamashita T, et al. Association of interleukin-28B genotype and hepatocellular carcinoma recurrence in patients with chronic hepatitis C. Clin Cancer Res. 2013;19:1827–37.
Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–11.
Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55.
Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–66.
Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406.
Chen X, Sun Y, Cai R, Wang G, Shu X, Pang W. Long noncoding RNA: multiple players in gene expression. BMB Rep. 2018;51:280–9.
Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46:2169–84.
He RZ, Luo DX, Mo YY. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6:6–15.
Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18.
Finci LI, Zhang X, Huang X, Zhou Q, Tsai J, Teng T, et al. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 2018;32:309–20.
Grzenda A, Lomberk G, Svingen P, Mathison A, Calvo E, Iovanna J, et al. Functional characterization of EZH2beta reveals the increased complexity of EZH2 isoforms involved in the regulation of mammalian gene expression. Epigenetics Chromatin. 2013;6:3.
Mu W, Starmer J, Yee D, Magnuson T. EZH2 variants differentially regulate polycomb repressive complex 2 in histone methylation and cell differentiation. Epigenetics Chromatin. 2018;11:71.
Chen K, Xiao H, Zeng J, Yu G, Zhou H, Huang C, et al. Alternative Splicing of EZH2 pre-mRNA by SF3B3 Contributes to the Tumorigenic Potential of Renal Cancer. Clin Cancer Res. 2017;23:3428–41.
Gao SB, Zheng QF, Xu B, Pan CB, Li KL, Zhao Y, et al. EZH2 represses target genes through H3K27-dependent and H3K27-independent mechanisms in hepatocellular carcinoma. Mol Cancer Res. 2014;12:1388–97.
Chen RY, Yen CJ, Liu YW, Guo CG, Weng CY, Lai CH. et al. CPAP promotes angiogenesis and metastasis by enhancing STAT3 activity. Cell Death Differ. 2020;27:1259–73.
Wang Y, Xu Y, Yan W, Han P, Liu J, Gong J, et al. CFIm25 inhibits hepatocellular carcinoma metastasis by suppressing the p38 and JNK/c-Jun signaling pathways. Oncotarget. 2018;9:11783–93.
Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65:798–808.
Wang J, Tai G. Role of C-Jun N-terminal kinase in hepatocellular carcinoma development. Target Oncol. 2016;11:723–38.
Zong X, Wang W, Ozes A, Fang F, Sandusky GE, Nephew KP. EZH2-mediated downregulation of the tumor suppressor DAB2IP maintains ovarian cancer stem cells. Cancer Res. 2020;80:4371–85.
Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–19.
Wang CH, Tsao YP, Chen HJ, Chen HL, Wang HW, Chen SL. Transcriptional repression of p21((Waf1/Cip1/Sdi1)) gene by c-jun through Sp1 site. Biochem Biophys Res Commun. 2000;270:303–10.
Ivanov VN, Bhoumik A, Krasilnikov M, Raz R, Owen-Schaub LB, Levy D, et al. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell. 2001;7:517–28.
Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene 2001;20:2390–400.
Doria M, Klein N, Lucito R, Schneider RJ. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–57.
Cougot D, Wu Y, Cairo S, Caramel J, Renard CA, Levy L, et al. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem. 2007;282:4277–87.
Qian Y, Wang B, Ma A, Zhang L, Xu G, Ding Q, et al. USP16 Downregulation by Carboxyl-terminal Truncated HBx Promotes the Growth of Hepatocellular Carcinoma Cells. Sci Rep. 2016;6:33039.
Yip WK, Cheng AS, Zhu R, Lung RW, Tsang DP, Lau SS, et al. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS One. 2011;6:e22888.
Iyer S, Groopman JD. Interaction of mutant hepatitis B X protein with p53 tumor suppressor protein affects both transcription and cell survival. Mol Carcinog. 2011;50:972–80.
Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, et al. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci USA. 1997;94:14707–12.
Lee S, Kim W, Ko C, Ryu WS. Hepatitis B virus X protein enhances Myc stability by inhibiting SCF(Skp2) ubiquitin E3 ligase-mediated Myc ubiquitination and contributes to oncogenesis. Oncogene. 2016;35:1857–67.
Srisuttee R, Koh SS, Kim SJ, Malilas W, Boonying W, Cho IR, et al. Hepatitis B virus X (HBX) protein upregulates beta-catenin in a human hepatic cell line by sequestering SIRT1 deacetylase. Oncol Rep. 2012;28:276–82.
Mai H, Zhou B, Liu L, Yang F, Conran C, Ji Y, et al. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:198.
El Marabti E, Younis I. The cancer spliceome: reprograming of alternative splicing in cancer. Front Mol Biosci. 2018;5:80.
Di C, Syafrizayanti, Zhang Q, Chen Y, Wang Y, Zhang X, et al. Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ. 2019;26:1181–94.
Luo ZL, Cheng SQ, Shi J, Zhang HL, Zhang CZ, Chen HY, et al. A splicing variant of Merlin promotes metastasis in hepatocellular carcinoma. Nat Commun. 2015;6:8457.
Li S, Hu Z, Zhao Y, Huang S, He X. Transcriptome-wide analysis reveals the landscape of aberrant alternative splicing events in liver cancer. Hepatology 2019;69:359–75.
Acknowledgements
We would like to thank Taiwan Liver Cancer Network (TLCN) for providing the hepatoma tissue samples and related clinical data (all are anonymous) for our research work.
Funding
This work was supported by grants from Chang Gung Memorial Hospital, Taoyuan, Taiwan (CMRPD1H0631-0633, CMRPD1K0291, NMRPD1G0941-0943, and NMRPD1G0951-0953 to K-HL; CMRPG3H0721-0723, NZRPG3G0171-0173, NMRPG3H0561, NRRPG3J0141 to Y-HL) and from the Ministry of Science and Technology of the Republic of China (MOST 106-2320-B-182-031-MY3; 106-2320-B-182-032-MY3; and 109-2320-B-182-011 to K-HL; MOST 106-2321-B-182A-004-MY3, MOST 107-2320-B-182A-028-, MOST 108-2320-B-182A-004- to Y-HL).
Author information
Authors and Affiliations
Contributions
Conception and design: Y-HL and M-HW. Data acquisition: Y-HL, M-HW, Y-CL, and P-CL. Data analyses and interpretation of data: Y-HL, M-HW, Y-CL, P-CL, C-TY, and K-HL. Drafting of the manuscript: Y-HL and M-HW. Revising critically the manuscript: C-TY and K-HL. Approval of final manuscript: Y-HL, M-HW, C-TY, and K-HL.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lin, YH., Wu, MH., Liu, YC. et al. LINC01348 suppresses hepatocellular carcinoma metastasis through inhibition of SF3B3-mediated EZH2 pre-mRNA splicing. Oncogene 40, 4675–4685 (2021). https://doi.org/10.1038/s41388-021-01905-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01905-3
This article is cited by
-
Novel long noncoding RNA LINC02820 augments TNF signaling pathway to remodel cytoskeleton and potentiate metastasis in esophageal squamous cell carcinoma
Cancer Gene Therapy (2023)
-
Decoding the role of aberrant RNA alternative splicing in hepatocellular carcinoma: a comprehensive review
Journal of Cancer Research and Clinical Oncology (2023)
-
CMAHP promotes metastasis by reducing ubiquitination of Snail and inducing angiogenesis via GM-CSF overexpression in gastric cancer
Oncogene (2022)
-
LINC00924-induced fatty acid metabolic reprogramming facilitates gastric cancer peritoneal metastasis via hnRNPC-regulated alternative splicing of Mnk2
Cell Death & Disease (2022)
-
Long non-coding RNAs are involved in alternative splicing and promote cancer progression
British Journal of Cancer (2022)