Abstract
Communication between acute myeloid leukemia (AML) and the bone marrow microenvironment is known to control disease progression. Therefore, regulation of AML cell trafficking and adhesion to the bone marrow is of significant interest. In this study, we demonstrate that differential expression of the membrane scaffold CD82 modulates the bone marrow homing of AML cells. By combining mutational analysis and super-resolution imaging, we identify membrane protein clustering by CD82 as a regulator of AML cell adhesion and bone marrow homing. Cluster analysis of super-resolution data indicates that N-linked glycosylation and palmitoylation of CD82 are both critical modifications that control the microdomain organization of CD82 as well as the nanoscale clustering of associated adhesion protein, N-cadherin. We demonstrate that the inhibition of CD82 glycosylation increases the molecular packing of N-cadherin and promotes the bone marrow homing of AML cells. In contrast, we find that the inhibition of CD82 palmitoylation disrupts the formation and organization of N-cadherin clusters and significantly diminishes bone marrow trafficking of AML. Taken together, these data establish a mechanism where the membrane organization of CD82, through specific posttranslational modifications, regulates N-cadherin clustering and membrane density, which impacts the in vivo trafficking of AML cells. As such, these observations provide an alternative model for targeting AML where modulation of protein organization within the membrane may be an effective treatment therapy to disrupt the bone marrow homing potential of AML cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Machida U, Kami M, Hirai H . Hematopoietic stem-cell transplantation for acute leukemia. N Engl J Med 1999; 340: 810; author reply 1-2.
Guzman ML, Allan JN . Concise review: Leukemia stem cells in personalized medicine. Stem Cells 2014; 32: 844–851.
Konopleva M, Konoplev S, Hu W, Zaritskey AY, Afanasiev BV, Andreeff M . Stromal cells prevent apoptosis of AML cells by up-regulation of anti-apoptotic proteins. Leukemia 2002; 16: 1713–1724.
Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE . Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med 2006; 12: 1167–1745.
Gibson LF . Survival of B lineage leukemic cells: signals from the bone marrow microenvironment. Leuk Lymphoma 2002; 43: 19–276.
Bradstock KF, Gottlieb DJ . Interaction of acute leukemia cells with the bone marrow microenvironment: implications for control of minimal residual disease. Leuk Lymphoma 1995; 18: 1–16.
Zhang B, Li M, McDonald T, Holyoake TL, Moon RT, Campana D et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-beta-catenin signaling. Blood 2013; 121: 1824–1838.
Bonardi F, Fusetti F, Deelen P, van Gosliga D, Vellenga E, Schuringa JJ . A proteomics and transcriptomics approach to identify leukemic stem cell (LSC) markers. Mol Cell Proteomics 2013; 12: 626–637.
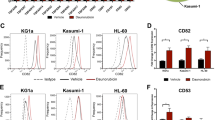
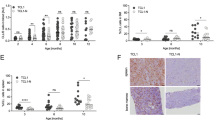
Nishioka C, Ikezoe T, Furihata M, Yang J, Serada S, Naka T et al. CD34(+)/CD38(-) acute myelogenous leukemia cells aberrantly express CD82 which regulates adhesion and survival of leukemia stem cells. Int J Cancer 2013; 132: 2006–2019.
Gil ML, Vita N, Lebel-Binay S, Miloux B, Chalon P, Kaghad M et al. A member of the tetra spans transmembrane protein superfamily is recognized by a monoclonal antibody raised against an HLA class I-deficient, lymphokine-activated killer-susceptible, B lymphocyte line. Cloning and preliminary functional studies. J Immunol 1992; 148: 2826–2833.
Imai T, Fukudome K, Takagi S, Nagira M, Furuse M, Fukuhara N et al. C33 antigen recognized by monoclonal antibodies inhibitory to human T cell leukemia virus type 1-induced syncytium formation is a member of a new family of transmembrane proteins including CD9, CD37, CD53, and CD63. J Immunol 1992; 149: 2879–2886.
Lebel-Binay S, Gil ML, Lagaudriere C, Miloux B, Marchiol-Fournigault C, Quillet-Mary A et al. Further characterization of CD82/IA4 antigen (type III surface protein): an activation/differentiation marker of mononuclear cells. Cell Immunol 1994; 154: 468–483.
Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995; 268: 884–886.
Boucheix C, Rubinstein E . Tetraspanins. Cell Mol Life Sci 2001; 58: 1189–1205.
Hemler ME . Targeting of tetraspanin proteins—potential benefits and strategies. Nat Rev Drug Discov 2008; 7: 747–758.
Bassani S, Cingolani LA . Tetraspanins: Interactions and interplay with integrins. Int J Biochem Cell Biol 2012; 44: 703–708.
Larochelle A, Gillette JM, Desmond R, Ichwan B, Cantilena A, Cerf A et al. Bone marrow homing and engraftment of human hematopoietic stem and progenitor cells is mediated by a polarized membrane domain. Blood 2012; 119: 1848–1855.
Takeichi M . Morphogenetic roles of classic cadherins. Curr Opin Cell Biol 1995; 7: 619–627.
Kemler R . From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet 1993; 9: 317–321.
Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003; 425: 841–846.
Bromberg O, Frisch BJ, Weber JM, Porter RL, Civitelli R, Calvi LM . Osteoblastic N-cadherin is not required for microenvironmental support and regulation of hematopoietic stem and progenitor cells. Blood 2012; 120: 303–313.
Greenbaum AM, Revollo LD, Woloszynek JR, Civitelli R, Link DC . N-cadherin in osteolineage cells is not required for maintenance of hematopoietic stem cells. Blood 2012; 120: 295–302.
Zhi L, Wang M, Rao Q, Yu F, Mi Y, Wang J . Enrichment of N-Cadherin and Tie2-bearing CD34+/CD38-/CD123+ leukemic stem cells by chemotherapy-resistance. Cancer Lett 2010; 296: 65–73.
Qiu S, Jia Y, Xing H, Yu T, Yu J, Yu P et al. N-Cadherin and Tie2 positive CD34(+)CD38(-)CD123(+) leukemic stem cell populations can develop acute myeloid leukemia more effectively in NOD/SCID mice. Leuk Res 2014; 38: 632–637.
Hong S, Troyanovsky RB, Troyanovsky SM . Binding to F-actin guides cadherin cluster assembly, stability, and movement. J Cell Biol 2013; 201: 131–143.
Nelson WJ . Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem Soc Trans 2008; 36: 149–155.
Termini CM, Cotter ML, Marjon KD, Buranda T, Lidke KA, Gillette JM . The membrane scaffold CD82 regulates cell adhesion by altering alpha4 integrin stability and molecular density. Mol Biol Cell 2014; 25: 1560–1573.
Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC . The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J Exp Med 1997; 185: 111–120.
Zaitseva L, Murray MY, Shafat MS, Lawes MJ, MacEwan DJ, Bowles KM et al. Ibrutinib inhibits SDF1/CXCR4 mediated migration in AML. Oncotarget 2014; 5: 9930–9938.
Yoshida T, Kawano Y, Sato K, Ando Y, Aoki J, Miura Y et al. A CD63 mutant inhibits T-cell tropic human immunodeficiency virus type 1 entry by disrupting CXCR4 trafficking to the plasma membrane. Traffic 2008; 9: 540–558.
Berditchevski F, Odintsova E, Sawada S, Gilbert E . Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J Biol Chem 2002; 277: 36991–37000.
Yang X, Kovalenko OV, Tang W, Claas C, Stipp CS, Hemler ME . Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J Cell Biol 2004; 167: 1231–1240.
Zhou B, Liu L, Reddivari M, Zhang XA . The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer Res 2004; 64: 7455–7463.
Dwivedi C, Dixit M, Hardy RE . Plasma sialyltransferase as a tumor marker. Cancer Detect Prev 1988; 11: 191–196.
Swindall AF, Londono-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL . ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res 2013; 73: 2368–2378.
Seales EC, Jurado GA, Singhal A, Bellis SL . Ras oncogene directs expression of a differentially sialylated, functionally altered beta1 integrin. Oncogene 2003; 22: 7137–7145.
Wang H, Zhang W, Zhao J, Zhang L, Liu M, Yan G et al. N-Glycosylation pattern of recombinant human CD82 (KAI1), a tumor-associated membrane protein. J Proteomics 2012; 75: 1375–1385.
White A, Lamb PW, Barrett JC . Frequent downregulation of the KAI1(CD82) metastasis suppressor protein in human cancer cell lines. Oncogene 1998; 16: 3143–3149.
Mazurov D, Heidecker G, Derse D . The inner loop of tetraspanins CD82 and CD81 mediates interactions with human T cell lymphotrophic virus type 1 Gag protein. J Biol Chem 2007; 282: 3896–3903.
Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl 2008; 47: 6172–6176.
Zhang J, Leiderman K, Pfeiffer JR, Wilson BS, Oliver JM, Steinberg SL . Characterizing the topography of membrane receptors and signaling molecules from spatial patterns obtained using nanometer-scale electron-dense probes and electron microscopy. Micron 2006; 37: 14–34.
Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL et al. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity 2013; 38: 461–474.
Ester M, Kreigel H, Sander J, Xu X . A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of 2nd International Conference on Knowledge Discovery and Data Mining 1996.
Chitaev NA, Troyanovsky SM . Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J Cell Biol 1998; 142: 837–846.
Xu Y, Zhuo J, Duan Y, Shi B, Chen X, Zhang X et al. Construction of protein profile classification model and screening of proteomic signature of acute leukemia. Int J Clin Exp Pathol 2014; 7: 5569–5581.
Malik FA, Sanders AJ, Jiang WG . KAI-1/CD82, the molecule and clinical implication in cancer and cancer metastasis. Histol Histopathol 2009; 24: 519–530.
Miranti CK . Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cell Signal 2009; 21: 196–211.
Huang F, Schwartz SL, Byars JM, Lidke KA . Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed Opt Express 2011; 2: 1377–1393.
Acknowledgements
We would like to acknowledge all of the funding sources that made this work possible. This includes funding from an NIH R01 HL122483-01A1 to JMG, a New Mexico INBRE grant subaward to JMG (NIH P20 GM103451), pilot funding from the University of New Mexico Cancer Center (NIH P30CA118100), an American Cancer Society Institutional Research Grant, a Post-Doctoral Training Fellowship to KDM (NIH T32 HL007736), Graduate Student Training Fellowships to CMT from the NM Spatiotemporal Modeling Center (NIH P50 GM085273) and (NIH F31 HL124977), and a Graduate Student Training Fellowship to CSR (NIH T32 HL007736). We also thank the NM Spatiotemporal Modeling Center for supporting the Super-Resolution Imaging Core Facility (NIH P50 GM085273) and the University of New Mexico Cancer Center (NIH P30CA118100) for use of the Keck-UNM Small Animal Models and Imaging Shared Resource. This work was also supported through faculty start-up funds to JMG from the University of New Mexico Department of Pathology. Finally, we would like to acknowledge the technical assistance of Rebecca J. Dodd and Dr I-Ming Chen, University of New Mexico Health Sciences Center. In addition, we would like to acknowledge the assistance of Dr Ravi Majeti, Division of Hematology, Institute for Stem Cell Biology and Regenerative Medicine, and Cancer Institute, Stanford University and the Stanford University Division of Hematology Tissue Bank for samples. (http://www.nature.com/onc)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Marjon, K., Termini, C., Karlen, K. et al. Tetraspanin CD82 regulates bone marrow homing of acute myeloid leukemia by modulating the molecular organization of N-cadherin. Oncogene 35, 4132–4140 (2016). https://doi.org/10.1038/onc.2015.449
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.449
This article is cited by
-
Microscopic clusters feature the composition of biochemical tetraspanin-assemblies and constitute building-blocks of tetraspanin enriched domains
Scientific Reports (2024)
-
Transcriptionally imprinted glycomic signatures of acute myeloid leukemia
Cell & Bioscience (2023)
-
Loss of bisecting GlcNAcylation on MCAM of bone marrow stoma determined pro-tumoral niche in MDS/AML
Leukemia (2023)
-
A KDM4A-PAF1-mediated epigenomic network is essential for acute myeloid leukemia cell self-renewal and survival
Cell Death & Disease (2021)
-
Tetraspanin CD82 drives acute myeloid leukemia chemoresistance by modulating protein kinase C alpha and β1 integrin activation
Oncogene (2020)