Abstract
Heat-shock factors (HSFs) are key transcriptional regulators in cell survival. Although HSF1 has been identified as a driver of carcinogenesis, HSF2 has not been explored in malignancies. Here, we report that HSF2 suppresses tumor invasion of prostate cancer (PrCa). In three-dimensional organotypic cultures and the in vivo xenograft chorioallantoic membrane model HSF2 knockdown perturbs organoid differentiation and promotes invasiveness. Gene expression profiling together with functional studies demonstrated that the molecular mechanism underlying the effect on tumor progression originates from HSF2 steering the switch between acinar morphogenesis and invasion. This is achieved by the regulation of genes connected to, for example, GTPase activity, cell adhesion, extracellular matrix and actin cytoskeleton dynamics. Importantly, low HSF2 expression correlates with high Gleason score, metastasis and poor survival of PrCa patients, highlighting the clinical relevance of our findings. Finally, the study was expanded beyond PrCa, revealing that the expression of HSF2 is decreased in a wide range of cancer types. This study provides the first evidence for HSF2 acting as a suppressor of invasion in human malignancies.
Similar content being viewed by others
Introduction
Prostate cancer (PrCa) is the most commonly diagnosed male cancer in Western countries.1 Gleason grading, which is based on glandular differentiation patterns within tumor biopsies, remains the standard for assessing prognosis and treatment.2 Although primary PrCa is often indolent, advanced PrCa can metastasize both locally and distantly. As PrCa progresses, it becomes resistant to pharmacological and surgical treatments and castration-resistant PrCa develops,1 which remains lethal.
The aggressiveness of cancer is connected to its invasive properties, which are governed by signaling pathways regulating dynamics of the cytoskeleton and turnover of cell–matrix and cell–cell junctions.3 The dynamics of cytoskeletal microfilaments is directed by plasma membrane receptors, including receptor tyrosine kinases, G-protein coupled receptors, integrins, transforming growth factor-β receptors, E-cadherins and Frizzled proteins. The cognate receptors activate members of the RHO GTPase family, which modulate effector protein activity.4 In PrCa studies, G-protein signaling and downstream pathway dynamics coincide with actin cytoskeleton reorganization, formation of invadopodia, extracellular matrix (ECM) degradation, modulations in adhesion and collagen contraction.4, 5, 6, 7 Molecular characterization of the transition from stable acinar morphology to local invasion, that is, the invasive switch, is inconclusive but needed as it may provide means to predict PrCa progression and facilitate discovery of treatments.
Heat-shock factors (HSFs) are multifaceted transcription factors that regulate the response upon proteotoxic stress, that is, the heat-shock response. HSF1 is the master regulator of stress responses and its targets include molecular chaperones, which maintain protein homeostasis.8, 9 HSF1 also regulates genes involved in, for example, cell cycle, protein synthesis, ribosome biogenesis and glucose metabolism.10, 11, 12 Importantly, HSF1 promotes cancer cell survival.10, 13, 14 The clinical significance of HSF1 was highlighted in a cohort of breast cancer patients, showing correlation between high HSF1 expression and decreased survival.15 Recently, HSF1-regulated transcriptional programs in malignant cells of several cancer types, and in tumor stroma, were reported to be different from the classical stress response.16, 17
In contrast to HSF1, another HSF family member HSF2, which can modulate HSF1-mediated stress responses,12, 18, 19, 20, 21 has hitherto not been associated with cancer. Genome-wide analyses revealed that HSF2 has an active role during mitosis, as it binds numerous loci in the human genome, despite global repression of the chromatin environment.12 Furthermore, decreased HSF2 expression during mitosis was shown to protect cells against proteotoxicity and apoptosis.22 Several cell lines, predominantly of tumorigenic origin, downregulate HSF2 expression during mitosis, perhaps correlating with the elevated levels of proteotoxic stress that cancer cells are subjected to,22, 23 thereby giving the cells a survival advantage.
Here, we demonstrate that HSF2 acts as a potent suppressor of tumor progression and invasion in PrCa by regulating signaling pathways steering epithelial plasticity. Furthermore, results from patient material imply functions of HSF2 in several human malignancies. Our findings strongly connect HSF2 to cancer and invasion, and advocate the use of HSF2-mediated regulation in therapeutics.
Results
Decreased HSF2 expression corresponds to high Gleason score and metastasis in PrCa patient samples
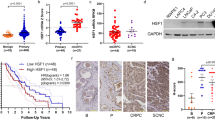
To explore a role of HSF2 in human malignancy, we analyzed HSF2 mRNA expression in a transcriptomic data set from 216 clinical PrCa samples.24 Interestingly, reduced HSF2 expression was found in PrCa samples compared with normal samples, and the expression was further decreased in metastatic samples (Figure 1a). At closer examination, low HSF2 expression correlated with high Gleason score tumors (Gleason scores ⩾8), which are prone to progress to castration-resistant PrCa and form distant metastases, as well as are associated with therapy failure and overall poor patient outcome.1 Concurrently, low HSF2 expression was associated with lymph node invasion, positive surgical margins and positive seminal vesicle invasion (Figure 1a), all indicating suppressive capacity of HSF2 on invasion/metastasis.
Low HSF2 expression correlates with aggressiveness of PrCa in human clinical samples. (a) Decreased HSF2 expression correlates with advanced PrCa. Analysis of mRNA expression for HSF2 in data sets from human PrCa (MSKCC),24 indicating that HSF2 expression is decreased in PrCa compared with normal tissue, and further decreased in advanced PrCa (Gleason score ⩾8), metastases, lymph node invasion (LN inv abnormal), positive surgical margins (SM) and positive seminal vesicle invasion (SV inv). (b) Oncoprints of HSF2, PTEN, TP53, MYC and HSF1 across 85 PrCa tumors,24 indicating genomic alterations. Individual genes are represented in each row, and individual tumors are represented as columns. Genetic alterations are color coded: dark blue, homozygous deletion; red, amplification; light blue frame, mRNA downregulation; pink frame, mRNA upregulation; green square, mutation. The oncoprints are based on data obtained from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal/). Z-score threshold±2. (c) Box and whisker plots indicating the expression of HSF2, PTEN or HSF1 mRNA across different subsets of PrCa tumors divided by the putative DNA copy-number status. Homdel, homozygous deletion; hetloss, heterozygous deletion; diploid, no change; gain, moderate amplification; amp, high-level amplification. HSF2 mRNA expression is as expected progressively lower across the different subgroups of PrCa with HSF2 ploidy status ranging from gain to homozygous deletion. Note that the scales for copy-number status (x axis) and mRNA expression (y axis) differ between the three graphs. The mRNA z-scores are compared with the expression in normal prostate samples. RAE, RAE algorithm.
The relevance of altered HSF2 expression was elucidated by comparing the genomic profile with that of known tumor suppressors or oncogenes in 85 PrCa samples displaying complete mRNA, copy-number and sequencing data.24, 25, 26 HSF2 was deregulated in 20% of the PrCa tumors, of which the majority (88%) showed a reduction in HSF2 expression level (Figure 1b and Supplementary Figure S1a). The expression of the well-known tumor suppressors PTEN and p53 was altered to a comparable degree as HSF2 in the same tumor set (18%, 29% and 20%, respectively). The expression of HSF1, another HSF family member and known transcriptional regulator in cancer cell survival,10, 13, 14, 15, 16 was also changed to the same extent as HSF2, although HSF1 mRNA was upregulated in the majority of cases (20%, Figure 1b).
Assessing putative copy-number alterations of HSF2, the reduction in HSF2 expression was mostly associated with heterozygous loss of HSF2 (Figure 1c). As a comparison, the relationship between expression and copy-number alterations for PTEN was typically associated with both heterozygous and homozygous deletions. HSF1, showing increased mRNA expression (Figure 1b), exhibited a higher range ploidy status (gain, amplification) (Figure 1c). Owing to the putative role of HSF2 in suppressing metastatic behavior, we assessed the clinical effect of deregulated expression levels in metastatic castrate PrCa samples.24 Although the cohort size was relatively small (23 metastatic biopsies), altered HSF2 expression correlated with poor disease-free survival (Supplementary Figure S1b). This validates the biological relevance of decreased expression observed in high-grade Gleason primary tumors.
HSF2 levels decline as PrCa cells advance towards an invasive phenotype and display dynamic expression during organotypic development
To investigate the molecular function of HSF2 in PrCa, we analyzed the mRNA expression in a panel of human prostate cell lines with different malignant potential and invasive behavior.5 HSF2 levels were very low in basal and non-transformed prostate epithelial cells (PrEC and EP156T) and high in malignant PrCa cell lines of the luminal type (LNCaP and Du145) (Figure 2a). Intriguingly, HSF2 expression was markedly lower in invasive cancer cells compared with that in less aggressive cells when comparing LNCaP and Du145 with PC3 cells, or PC3 cells with the more aggressive PC3-M cells (Figure 2a). This pattern of expression was concomitant with the divergent HSF2 protein levels observed in basal, non-transformed EP156T, malignant but noninvasive luminal LAPC4 and highly invasive PC3 cells (Figure 2b). In contrast, the protein levels of HSF1 did not decrease in the invasive PC3 cells (Figure 2b).
Expression patterns of HSF2 in PrCa cell lines and during organotypic PrCa development. (a) mRNA expression of HSF2 in basal, non-transformed prostate epithelial cells (PrEC, EP156T), luminal noninvasive PrCa cell lines (LNCaP, Du145) and luminal invasive PrCa cell lines (PC3, PC3-M), derived from Illumina arrays.5 The results are from two individual experiments. Error bars indicate s.e.m. (b) Western blot analysis of HSF2 and HSF1 protein expression in basal, non-transformed prostate epithelial cells (EP156T), PrCa cells of a noninvasive type (LAPC4) and PrCa cells of an invasive type (PC3). Hsc70 was used as a loading control. (c) Schematic illustration of PrCa cells grown as 3D culture in the Matrigel or as 2D monolayer culture on plastic. The former allows organotypic organoid formation, acinar differentiation and invasion. The basement membrane is depicted in blue. (d) HSF2 mRNA expression analysis of PC3 cells grown in 2D or allowed to differentiate in 3D for the indicated days (d4, d8, d13 and d15). The results are from two individual experiments. Error bars represent ±s.e.m. (e) Western blot analysis of HSF2 and HSF1 protein expression in PC3 cells grown in 2D or allowed to differentiate for the indicated days in 3D organotypic culture. Tubulin was used as a loading control. inv, Invasive; non-inv, noninvasive.
Next, the expression pattern of HSF2 was examined in three-dimensional (3D) organotypic cell cultures. The 3D model promotes organotypic acinar structures, that is, organoids, which display physiologically relevant cell–cell and cell–matrix interactions, epithelial polarization and differentiation, thus recapitulating human cancer histology in vivo (Figure 2c).5, 27, 28 In 3D ECM culture, tumor-derived PC3 cells transiently differentiate into hollow organoids around days 4 and 5, but spontaneously dedifferentiate (around days 8–10) into invasive stellate structures (days 11–13; Figure 2c).5 HSF2 mRNA expression peaked during spheroid polarization and differentiation, and decreased simultaneously with the appearance of invasive structures (Figure 2d). At the protein level, this pattern was even more pronounced, and in stark contrast to the uniform expression of HSF1 (Figure 2e). Taken together, these results suggest that HSF2 maintains the luminal phenotype and promotes epithelial maturation.
HSF2 suppresses invasive behavior of PrCa organoids
To study the functional consequences of the dynamic expression of HSF2 in tumor progression, HSF2 was silenced in 3D organotypic cell culture models. PC3 cells were chosen as they are widely used as an experimental model for castration-resistant PrCa and metastasis. We monitored the effects on morphology, polarization and growth of PrCa cells, which possess both a striking differentiation capacity into organoids (acini), but that can also acquire massive invasive properties, characteristic of aggressive cancer cells. Cells were transfected with small interfering RNAs (siRNAs) (Figure 3a), transferred into the Matrigel and organoid morphology was monitored for 8 days. Cells transfected with control siRNA initially matured into well-differentiated organoids and spontaneously formed invasive, multicellular structures around day 8, followed by overt invasion shortly thereafter (Figure 3b and Supplementary Video S1a). Interestingly, HSF2 silencing further increased the invasive properties, and the onset of invasion was detected at an earlier time point (Figure 3b and Supplementary Video S1b). In contrast, HSF1 silencing, used as a control, perturbed acinar differentiation and increased the frequency of cell death, thus prohibiting subsequent formation of invasive structures (Figure 3b and Supplementary Video S1c). It is notable that silencing of HSF2 or HSF1 did not markedly affect cell proliferation or morphology, as measured in two-dimensional (2D) cultures (Supplementary Figure S2).
Loss of HSF2 promotes PrCa organoid invasion. (a) Western blot of the silencing efficacy in PC3 cells 48 h after transfection with siRNAs against a scrambled sequence (Ctrl), HSF1 or HSF2. Actin was used as a loading control. (b) Live-cell imaging of invasive PC3 cells transfected with the indicated siRNAs and grown in 3D culture. Representative images from days 1 to 8 in 3D culture are shown. Scale bar, 100 μm. (c) Confocal laser scanning microscopy images of PC3 cells grown in 3D for 2, 5, or 8 days after transfection with siRNAs. Alexa488-labeled phalloidin was used to detect filamentous actin (F-actin; green) and DAPI (4',6-diamidino-2-phenylindole) to label nuclei (blue). Scale bar, 50 μm. (d) Representative segmentation of live-cell spinning disk confocal images of PC3 organoids transfected with siRNAs as in panel a, and grown for 8 days in 3D culture. Organoids were segmented and analyzed by the AMIDA.29 Apoptotic cells are displayed in red (Nucview) and live cells in green (SYTO62). (e) Upper panel: heatmap and graphical P-value matrix, summarizing the altered morphometric parameters from the AMIDA analysis. The total number of organoids measured were 1650 (Ctrl), 1134 (HSF1 siRNA) and 1791 (HSF2 siRNA). Values are color coded as red (increased) and blue (decreased), in response to siRNA knockdown of HSF1 and HSF2, relative to control. P-values are Bonferroni corrected from t-tests and compared with scrambled control. For explanations of morphometric parameters, see Supplementary Table 1. Lower panel: box and whisker plots of selected parameters from the upper panel. Roundness is a measure of loss of the round organoid phenotype (Roundness), and is associated with invasive properties, whereas roughness of the segmented structures (Roughness), and severity of cellular protrusions emerging from the core structure (AppIndex), both relate to local invasion and cell motility. P=0 indicates P-values <0.001.
We examined organoid morphology in more detail by staining nuclei and F-actin for confocal microscopy (Figure 3c). Starting around day 5, HSF2-depleted organoids already exhibited pronounced collective invasive behavior, whereas organoids with silenced HSF1 were smaller and remained round beyond day 8. The phenotypes of over thousand organoids were analyzed by the high-content, automated morphometric image data analysis software (AMIDA).29 AMIDA allows segmentation and quantitative measurements of large numbers of images containing hundreds of organoids, with different shapes, sizes and textures (Figure 3d). As summarized in Figure 3e, significant differences in many parameters were found between control organoids and those depleted of either HSF2 or HSF1 (Figure 3e (upper panel) and Supplementary Table 1). Silencing of HSF2 promoted invasion as it led to loss of the round shape of the organoids (Roundness), an increase in the number of small filopodia-like cellular extensions (Roughness) and in the severity (AppIndex) and length (MaxApp) of invasive multicellular structures (Figure 3e). Invasion was generally of the collective type, that is, as a string of connected cells, with only a few single, migrating cells. In contrast, HSF1-silenced organoids maintained their round shape, showed lower numbers of small filopodia and less invasive structures. The size of the organoids was decreased (Area) and apoptosis increased (AreaRatioR, R=red channel). In summary, these results indicate that the function of HSF2 is strikingly opposite to that of HSF1 in growth and differentiation of PrCa organoids.
To validate the function of HSF2 on organoid morphology, rescue experiments were performed in which HSF2 was silenced using a short hairpin RNA (shRNA)-producing construct as described previously.19 Thereafter, the expression of HSF2 was restored by overexpressing an RNA interference (RNAi)-resistant HSF2 construct. In accordance with the results obtained using siRNAs against HSF2 (Figure 3), a significant increase in invasiveness was detected after silencing HSF2 using the shRNA-producing plasmid (Figure 4). When the expression of HSF2 was restored, the biological effect was reversed (Figure 4), thereby demonstrating a function of HSF2 on invasiveness.
Rescue of HSF2 expression reverses the invasive phenotype of PrCa organoids. (a) Western blot of PC3 cells growing in 2D, 48 h after transfection. Cells were transfected with shRNA-producing plasmids against a scrambled sequence (Ctrl) or HSF2, and the HSF2 expression was restored by overexpression of an RNAi-resistant HSF2 construct (Mut HSF2). The exogenous HSF2 is marked with an asterisk. Tubulin was used as a loading control. (b) Live-cell imaging of invasive PC3 cells grown in 3D and transfected with the indicated shRNA constructs at day 3 in organotypic culture. Representative images from days 0, 3, 5 and 7 in 3D culture are shown. Scale bar, 100 μm. (c) Confocal microscopy images of immunofluorescence stainings in PC3 cells cultured in 3D for 8 days, and transfected as in (b). Upper panel: Alexa488-labeled phalloidin was used to detect filamentous actin (F-actin; green), and DAPI (4',6-diamidino-2-phenylindole) to label nuclei (blue). Lower panel: HSF2 staining was used to detect the silencing and rescue efficacy in PC3 organoids. No antibody staining (No Ab Ctrl) was used as a negative control. Scale bar, 50 μm.
HSF2 is a regulator of cell movement and invasion by affecting actin cytoskeleton signaling
The underlying mechanisms for the profound changes in tumor organoid phenotypes in differentiating PC3 organoids in the presence or absence of HSF2 were investigated. We silenced HSF2 using RNAi in PC3 cells, transferred them into 3D culture and performed gene expression profiling using whole-genome microarrays. The silencing efficacy was assayed at different time points (Supplementary Figure S3a). Gene expression was analyzed after 5 and 8 days in 3D culture, representing critical stages at which organoids either undergo acinar differentiation (day 5) or transform spontaneously into invasive structures (day 8).5 Significant up- and downregulation of genes were detected at both time points. The complete gene expression data set is available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; data set GSE48672).
Gene set enrichment analysis30 showed that genes affected by HSF2 silencing were strongly related to gene ontology (GO) terms associated with translation, energy metabolism, transmembrane transport, RNA or chromatin (Figure 5a and Supplementary Table 2). Further down in the list were GO terms such as mitosis, lipid homeostasis and metabolism (Supplementary Table 2). Most interestingly, HSF2 silencing affected numerous GO terms linked to invasion. At day 5, GTPase activity and GTP binding were among the top-ranked terms (Figure 5a, Supplementary Table 2 and Supplementary Figure S3b). Within these terms, 38 genes were differentially expressed upon HSF2 silencing (Figure 5b), including members of the Rho family of GTPases (e.g. RHOA, RND3) and stimulatory G proteins (e.g. GNA12/13, GNAQ). Other enriched, invasion-associated GO terms included collagen, adhesion and ECM (Figure 5a and Supplementary Table 2). Functional annotations on the top-ranked 300 genes investigated with the DAVID analysis tool31 showed strong deregulation of adhesion and ECM, including genes of the cadherin, integrin, laminin and collagen families (Figure 5c, left panel). At day 8, when organoids were transformed into the invasive morphology, GO terms downstream of GTPase activity, such as small GTPase-mediated signal transduction and actin cytoskeleton, showed gene enrichment (Figure 5a, Supplementary Table 2 and Supplementary Figure S3b). Among the genes were key GTPase effector proteins ROCK2, WASF2 and several filamins and myosins (Figures 5b and c, middle and right panels), which act as executors of actin cytoskeleton function.4, 32 Ingenuity pathway analysis highlighted how HSF2 silencing interfered with the actin cytoskeleton pathway at various levels, and that loss of HSF2 expression was prominently linked to increased cell movement and invasiveness (Figure 5d). Our findings are in accordance with the massive rearrangements of the actin cytoskeleton observed upon transformation of mature acini into invasive PrCa organoids5, 7 and with the phenotype upon HSF2 silencing (Figure 3).
Altered gene expression patterns in the absence of HSF2 in PrCa organoids. PC3 cells were transfected with scrambled control siRNA or siRNA against HSF2. Total RNA was purified at day 5 or 8 from triplicate cultures of PC3 organoids, RNA was hybridized to Illumina micorarrays and the relative gene expression was analyzed. (a) Gene set enrichment analysis (GSEA) of mRNA expression data of PC3 organoids at day 5 or 8 in 3D culture. Values indicate relative enrichment of gene sets after HSF2 silencing compared with control. The complete list of GO terms is shown in Supplementary Table 2. Enrichment plots for selected GO terms is shown in Supplementary Figure S3b. (b) Venn diagram of the overlap of genes related to G-protein signaling (GTPase activity, small GTPase-mediated signal transduction and regulation thereof) and actin cytoskeleton (actin cytoskeleton, actin filament-based processes, actin cytoskeleton organization and biogenesis and actin binding), differently expressed in PC3 organoids upon HSF2 silencing at day 5 or 8 in 3D culture. The number of altered genes, both pathway specific and shared between the pathways, upon HSF2 silencing, is indicated inside the circles. The figure was constructed using Biovenn bioinformatics tool.59 (c) Gene term heatmaps generated in the DAVID analysis tool from the top 300-ranked target genes. Focal adhesion (left panel) gene clusters were enriched at day 5 and RHO GTPase (middle panel) and actin cytoskeleton-related gene clusters (right panel) at day 8 upon HSF2 silencing. Green areas show common annotations between terms and genes. (d) Schematic illustration based on Ingenuity Pathway Analysis of genes in actin cytoskeleton signaling. HSF2 silencing interfered with the actin cytoskeleton signaling by regulating genes at various levels, as revealed by microarray analysis. Genes affected by HSF2 knockdown in invasive 3D structures at day 8 are shown in red (upregulated) and green (downregulated). The small molecular inhibitors used in Figure 6b are indicated in blue. Act, activity; dep, dependent; GO, GO terms; KEGG, kegg pathways, KEY, keywords, reg, regulation.
The above-mentioned results lay the ground for a hypothesis that HSF2 acts as a regulator of normal acinar morphology versus invasion. To test the hypothesis and to confirm the results of the microarray analyses, the link between HSF2 and pathways facilitating invasive properties, that is, GTPase activity and actin cytoskeleton signaling, was explored. First, we investigated GTPase regulation in PC3 cells in organotypic 3D culture, in the presence or absence of HSF2. Silencing of HSF2 resulted in a prominent increase in the protein level of the stimulatory G-protein guanine nucleotide-binding protein α13, GNA13 (Figure 6a), which is in line with previous reports on its elevated expression upon activation.33, 34 Concomitantly, the protein levels of Rho GTPase-activating protein 1, ARHGAP1, a converter of Rho, Rac and Cdc42 proteins to the inactive GDP-bound state,35 decreased. As expected, owing to regulation via GTP-GDP exchange,36 no significant changes were detected in the amount of Cdc42 (Figure 6a), which is the preferred target of ARHGAP1. Second, key modulators of the pathways facilitating invasive properties were specifically blocked and the resulting effects compared with those observed upon HSF2 silencing. Chemical inhibition of the RHO-associated protein kinase ROCK by Y27632 (Figure 5d) led to a more pronounced invasive phenotype, as detected by the formation of small filopodia, increase in the length of invasive structures and loss of the round organoid shape (Figures 6b and c and Supplementary Figure S4). This phenotype is concomitant with that observed previously in 3D culture,7, 29 and interestingly, with the phenotype detected upon HSF2 silencing (Figure 3). Rac downstream activity, which counteracts the RHOA-ROCK axis,32 was blocked by the PAK inhibitor IPA3. The treatment with IPA3 led to a complete block of invasiveness (Figures 6b and c and Supplementary Figure S4), in accordance with earlier results.29 Most importantly, the invasive phenotype induced by HSF2 silencing was effectively reversed by subsequent inhibition of RAC/PAK with IPA3 (Figures 6b and c and Supplementary Figure S4). These results support a role of HSF2 in PC3 organoids undergoing invasive transformation via GTPase signaling and regulation of actin cytoskeleton pathways.
HSF2 is linked to G-protein signaling, focal adhesion and actin cytoskeleton during PrCa progression. PC3 cells were transfected with siRNA against a scrambled sequence (Ctrl) or HSF2, and cultured in 3D. (a) Western blotting was performed with antibodies against GNA13 (G-protein guanine nucleotide-binding protein α13), ARHGAP1 and Cdc42 after 5 days in 3D culture. Hsc70 was used as a loading control. (b) PC3 cells were transfected and cultured as above and thereafter treated with DMSO, RAC-downstream PAK inhibitor (IPA3) or ROCK kinase inhibitor (Y26732), as indicated in the figure. Inhibitors were used at a concentration of 10 μM. Cells were stained by calcein AM and imaged by spinning disk confocal microscopy (x5) at day 7. Scale bar, 100 μm. The images are blow-ups from Supplementary Figure S4. (c) Box and whisker plots of image data from panel b measuring roughness of the segmented structures (Roughness), length of cellular protrusions emerging from the core structure (MaxApp) and loss of the round organoid phenotype (Roundness), all of which correlate with invasive properties. P=0 indicates P-values <0.001. (d) mRNA expression analysis of EMT markers CDH1, SNAI2, TWIST1, S100A4, FN1, VIM and CDH2 of PC3 cells grown in 3D in the presence (Ctrl) or absence of HSF2 (HSF2 siRNA). The results are from five independent experiments, and represent fold change normalized to the respective scrambled day 8 value. Error bars indicate s.e.m. *P-values <0.05; **P-values <0.01 by two-tailed paired Student's t-test. (e) PC3 cells were treated as in (d) and western blot analysis of EMT markers E-cadherin, Snail 2, vimentin and N-cadherin was performed. Hsc70 was used as a loading control.
Epithelial–mesenchymal transition (EMT) is a critical differentiation program by which epithelial cells lose their cell polarity and cell–cell adhesion, and simultaneously gain migratory and invasive properties to become mesenchymal-like cells that contribute to tumor progression.37, 38 To address if the observed effects of HSF2 silencing on invasion can be attributed, at least in part, to EMT, we examined the expression of a panel of EMT markers in organotypic culture of PC3 cells upon HSF2 knockdown. Silencing of HSF2 markedly reduced both mRNA and protein expression of E-cadherin (CDH1) (Figures 6d and e), which is considered a hallmark of EMT.39 The EMT-inducing transcription factors SNAI2 (Slug/Snail 2) and Twist (TWIST1), as well as mesenchymal markers fibroblast-specific protein 1 (S100A4), fibronectin (FN1) and vimentin (VIM) were correspondingly upregulated at mRNA or protein levels upon HSF2 silencing (Figures 6d and e). In addition, the protein level of N-cadherin (CDH2) increased at day 5 in the absence of HSF2 (Figure 6e). Taken together, these results suggest that HSF2 silencing promotes EMT and plasticity of PC3 cells, which may be associated with the invasive behavior of HSF2-depleted PrCa organoids.
Loss of HSF2 increases tumor growth and invasive properties in vivo
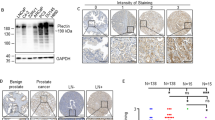
To validate the functional role of HSF2 in tumor formation, growth and invasion in vivo, we used the chorioallantoic membrane (CAM) model.40, 41 PC3 cells were transfected with shRNA-producing plasmids to abrogate HSF2 expression19 (Figure 7a), and grafted onto the exposed CAM of chicken embryos. HSF2 silencing resulted in significantly larger tumors formed by PC3 cells compared with controls, as evaluated by the tumor area (Figures 7b and c). Histological examination on hematoxylin–eosin (HE)-stained sections demonstrated that the implanted tumor cells, both non-transfected and control cells, developed into poorly differentiated adenocarcinomas (Figure 7d). Although HSF2 silencing resulted in adenocarcinomas of similar histology, single tumor cells seemed more often to escape the primary tumor site (Figure 7d). Owing to the reduced susceptibility of HSF1-knockout mice to tumor formation, we included HSF1 silencing in the analyses. Cells depleted of HSF1 formed only small tumors that were hardly detectable and highly fibrotic (Figures 7b–d). These results demonstrate strong tumor-promoting and -suppressive effect of HSF1 and HSF2, respectively.
Silencing of HSF2 increases in vivo tumor growth in the CAM model. PC3 cells were transfected with indicated shRNA constructs to silence either HSF2 or HSF1 and assayed 3 days after that 1 × 106 cells were implanted on CAM. (a) The protein expression of shRNA-transfected cells was examined 4 days after transfection by western blotting against HSF2 and HSF1. Tubulin was used as a loading control. (b) Images of representative tumors on the CAM. Upper panel: overview image of the control (Ctrl)-treated egg. Lower panel: the implantation area is confined by a plastic ring, inside of which the tumors are located. Scale bar, ~1 mm. (c) Relative tumor area was determined using ImageJ. The area of the control-transfected tumor (Ctrl) was arbitrarily set to 1. Results are from four independent experiments, each with 3–5 eggs per shRNA construct. Error bars indicate s.e.m. **P-values <0.05 by Student's t-test. (d) Representative images of HE-stained primary tumors either non-transfected or transfected with the indicated shRNA. HSF1 silencing caused almost complete abolishment of PC3 tumors. Cells escaping from the primary tumor site were most frequently detected after HSF2 depletion as indicated by arrows. The insets show blow-ups. Scale bar, 50 μm. (e) Representative immunohistochemistry (IHC) images of sections stained against human MMP14 after transfection with control (Ctrl) or HSF2-targeting shRNA constructs. In each image, both the PC3-derived tumor (T) and chicken-derived CAM (C) are shown. In the image pairs, the blow-ups (right) represent the framed area (left).
We investigated the invasive properties of HSF2-depleted tumors further by staining sections against MMP14, which is a membrane-type matrix metalloproteinase that facilitates local ECM degradation. In cancer, MMP14 is crucial for cell migration and invasion, and high expression levels correlate with advanced stages of PrCa.42, 43 Intriguingly, tumors where HSF2 was silenced exhibited strong MMP14 staining at the edge of the tumor mass, which was not the case in control-transfected tumors (Figure 7e). In addition, cellular localization of MMP14 was more frequently membrane tethered in HSF2-depleted cells compared with the diffuse cytoplasmic staining of control-transfected cells. The proteolytic activity of MMP14 involves its translocation to the cell surface, as seen in many neoplastic cells types including PC3 cells,42, 43 further supporting our finding that loss of HSF2 promotes tumor invasion.
Correlation between HSF2 expression and its target genes in clinical samples
As HSF2 affected expression of genes associated with invasion and migration (Figures 5 and 6), mRNA levels of a selection of putative HSF2 target genes related to GTPase activity, focal adhesion, small GTPase-mediated signaling and actin cytoskeleton were assessed in human clinical PrCa samples.24 Validating our data from 3D cultures, many of the target genes, such as ITGA5 (integrin α5), COL6A3 (collagen α6), FLNA (filamin A) and ROCK2 (RHO-associated protein kinase 2), showed mRNA expression pattern similar to that of HSF2 (Figure 1a), that is, reduced expression in tumors versus normal tissue and further decreased expression in high Gleason score tumors and metastatic samples (Figure 8a). When comparing expression levels in individual tumors, approximately half of the putative HSF2 target genes showed tendency toward co-occurrence with HSF2 (Figure 8b and Supplementary Figure S5). Altered expression levels of target genes in comparison with HSF2 in individual tumors were further visualized by oncoprints (Supplementary Figure S6). These results suggest that the function of HSF2 is directed via invasion-related genes also in clinical human PrCa.
Correlation between HSF2 expression and its target genes in clinical samples. (a) Analysis of mRNA expression in human PrCa samples24 of a selection of putative HSF2 target genes identified by the microarray analysis (Figure 5). The target genes follow the same expression trend as HSF2 (Figure 1a), that is, decreased levels in PrCa compared with normal tissue, and further decreased in advanced PrCa (Gleason score ⩾8) and metastases. (b) HSF2 target genes associated with the gene set enrichment analysis (GSEA) GO terms GTPase activity, focal adhesion, small GTPase-mediated signaling or actin cytoskeleton exhibiting co-occurrence (light or dark yellow) or mutual exclusivity (light or dark blue) with HSF2 when comparing individual tumors using the cBioPortal database. The complete target gene lists are shown in Supplementary Figure S5. P-values <0.05, as derived via Fisher's exact test, are outlined in red. P-values are not adjusted for false discovery rate.
HSF2 expression is decreased in several human malignancies
Finally, we expanded the study to human malignancies other than PrCa by assessing alteration frequency of HSF2 expression across a large collection of cohorts.44 Intriguingly, low levels of HSF2 mRNA were detected in many types of malignancies (Figure 9a), including other epithelial cancers, for example, invasive breast carcinoma, small-cell lung carcinoma and ovarian serous papillary carcinoma (Figures 9a and b, upper panel). In addition, non-epithelial cancers exhibited decreased HSF2 expression in cancerous tissue when compared with normal tissue, and the highest fold changes were detected in clear-cell renal cell carcinoma, embryonal carcinoma and glioblastoma (Figures 9a and b, lower panel). Thus, reduced HSF2 expression is not restricted to PrCa or other epithelial cancer types, but is found in a broad range of malignancies.
HSF2 expression is frequently decreased in several human malignancies. (a) HSF2 mRNA expression in cancer patient samples compared with control tissue analyzed by the Oncomine database (https://www.oncomine.org). The origins of the cohorts are displayed in the figure.60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75 Shown are fold change with a cutoff value of −2. (b) Expression levels (log 2) of HSF2 mRNA in individual patient samples of small-cell lung carcinoma vs lung, invasive breast carcinoma vs breast, clear-cell renal carcinoma vs kidney, embryonal carcinoma vs testis and glioblastoma vs neural stem cells. The cohorts are the same as in panel a. Upper panel: epithelial cancer types; lower panel: non-epithelial cancer types. Adenocarc, adenocarcinoma; Carc, carcinoma; FC, fold change; P, P-value.
Discussion
Unlike HSF1, which drives carcinogenesis, HSF2 had not been connected to cancer before this study, where we addressed the impact of HSF2 on PrCa. The expression of HSF2 varied in cell lines representing different malignant potential, as well as during maturation and spontaneous invasive transformation of PrCa organoids. HSF2 is a short-lived protein, whose activity is regulated via its abundance,20, 22, 45, 46, 47 and therefore, the expression dynamics detected here likely reflects the transactivating capacity of HSF2. HSF2 was abundant in transformed cells with weak invasive potential and during the acinar differentiation phase of PrCa organoids, whereas in invasive cell types and during dedifferentiation into invasive stellate organoids, HSF2 was present in minute amounts. Moreover, HSF2 depletion stimulated polarized epithelial PrCa cells to dedifferentiate and chose a more aggressive invasive path. HSF2 silencing specifically increased the collective invasion, which is characteristic for cancer cells in organotypic 3D cultures.48 These results suggest that HSF2 is involved in epithelial maturation and acts as a negative regulator of cancer cell invasion. We propose that HSF2 has an important role at the time of cell fate decision; determining whether the cells remain in a glandular-epithelial state of differentiation or progress through spontaneous transformation into highly invasive state. Thus, HSF2 would be a key determinant at the invasive switch.
How does HSF2 mediate its profound effects on PrCa progression? Gene profiling after silencing HSF2 in cancer organoids revealed strong enrichment of GO terms related to epithelial plasticity. In accordance, high degree of epithelial plasticity is displayed during spontaneous invasive transformation, as demonstrated by altered expression of gene sets related to cell adhesion, cell–cell contacts, invasion/metastasis and ECM turnover during PrCa organoids progression.5 The result also matches our finding that HSF2 silencing promotes earlier onset of the invasive switch and more pronounced invasive properties (Figure 3).
The 3D organotypic biomimetic platform allowed observations of the silencing effects specifically at a time when cells are differentiated (acinar stage), before the invasive switch (day 5), when spontaneous dedifferentiation starts (days 7 and 8), and when cells form overt, invasive structures (after day 8). At day 5, HSF2-affected gene sets were related to collagen and adhesion, which is coherent with the changes in collagen metabolism, the ECM and cell–cell contacts as early indicators of invasion.49, 50 In addition, HSF2 silencing led to deregulation of GTP binding and GTPase activity, including genes functioning as hubs in signaling from the microenvironment.4 Further investigations revealed that HSF2 downregulation also promoted tumor plasticity via EMT. At day 8, RHO GTPase and small GTPase signaling pathways were among the most enriched GO terms, both of which are linked to actin cytoskeleton dynamics, downstream of G-protein-mediated signaling. These results equal epithelial plasticity pathway kinetics and dynamics.4 On the gene level, the interconnection between HSF2-affected pathways was illustrated by a Venn diagram (Figure 5b), where the single common denominator between the GO terms was RHOA. The three genes deregulated at both days 5 and 8 were RHOD, RRAGC and GNB1. RHOA is a small GTPase that is primarily known as a central regulator of the actin cytoskeleton via its effector protein ROCK, and has been associated with essentially all stages of cancer progression.32 Previously, it was reported that the invasive switch of PC3 cells is linked to repression of Gα/RHOA/ROCK1 and 2 signaling, and that members of this pathway, for example, RHOA, ROCK and myosin, are downregulated in invasive PC3 cells.7 These results suggest that HSF2 promotes acinar morphology and represses the spontaneous invasive switch by interfering with genes important for, for example, adhesion, ECM, EMT and G-protein-coupled signaling. In the absence of HSF2, the effect, from deregulation of the above-mentioned pathways, is conveyed to downstream targets and detected by increased invasion.
The finding that HSF2 silencing led to increased tumor growth and invasion matches the low HSF2 levels observed in high Gleason score patient tumors and the decrease in metastatic samples. This indicates that low HSF2 levels permit invasive behavior and propose a role for HSF2 as a suppressor. A notorious tumor suppressor in PrCa is PTEN, for which heterozygous deletion correlates with early stage and indolent tumors, and homozygous deletion correlates with late-stage, aggressive PrCa.51 When comparing the mRNA expression levels of PTEN and HSF2, downregulation was detected in a similar percentage of tumors within the cohort (Figure 1b). Another tumor suppressor is NME1, which was identified based on its low expression in metastatic cells. Albeit NME1 was the first gene recognized as a metastasis suppressor, the mechanism underlying its metastatic potential remains controversial.52, 53 The metastasis suppressor CD82 is expressed at low levels in PrCa, which, together with loss of p53, announces poor patient survival.54, 55 Similarities between HSF2 and tumor/metastasis suppressors, in terms of prevalence and function, are obvious. A role of HSF2 as a suppressor is further supported by the finding that a large portion of HSF2 target genes in clinical samples are important for invasion and metastasis (Figure 8).
We propose a model in which HSF2 functions as a suppressor of EMT, tumor plasticity and invasion in tumorigenesis (Figure 10). Based on our results from 3D organotypic cultures combined with clinical patient material, the expression of HSF2 peaks in differentiated cells not yet having advanced through the invasive switch. Decreased expression levels of HSF2 are a determinant of PrCa progression, likely through deregulation of key infiltration-associated pathways, resulting from altered transactivation potential of HSF2.
Schematic model on how HSF2 expression levels may determine PrCa progression and invasion via interference with molecular signaling pathways. High levels of HSF2 are associated with a differentiated state of prostate cells and with normal tissue in patient samples. At this stage, HSF2 activity represses invasion-provoking pathways related to, for example, adhesion (ECM and collagen) and GTPase activity. In malignant cells, the expression level of HSF2 decreases with a concomitant disruption of HSF2-regulated pathways, leading to the formation of invasive structures. In highly invasive cells and metastatic tissues, a further decline in the expression level of HSF2 is apparent. The dynamic expression level of HSF2 is illustrated by the green triangle.
PrCa often remains indolent, but if the disease progresses into advanced stages, no cure is currently available.51 A therapeutic option is therefore inhibiting invasion when instigated. The results presented herein suggest that HSF2, together with associated gene signatures, could constitute such therapeutic targets. This is demonstrated in patients with metastatic tumors in which altered HSF2 expression correlates with poor disease-free survival (Supplementary Figure S1b). In-depth studies are warranted but are challenging to perform owing to the aggressiveness of the disease at this stage and consequential lack of samples. Furthermore, HSF2 is an upstream regulator of oncogenic mechanisms relevant for tumor progression and invasion, which are attractive therapeutic targets. Our study sets the stage for therapeutic opportunities for intervening with aggressive PrCa early in the signaling cascade leading to invasion. In addition, loss of HSF2 expression across many human malignancies indicates a fundamental role in tumorigenesis, which deserves future exploration.
Materials and methods
Bioinformatics of mRNA expression in clinical samples
Expression and correlation of HSF2 with other genes and pathways were analyzed in a clinical transcriptome study (Memorial Sloan Kettering Cancer Centre, MSKCC),24 available through the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal).25, 26 The collection contained 216 PrCa samples and metastases, of which 85 display complete mRNA, copy-number and sequencing data, based on Affymetrix Human Exon 1.0 ST Arrays and next-generation sequencing. Part of this data set, the metastatic castrate samples, was used for survival studies. Furthermore, gene expression data from curated and normalized values of the MSKCC collection was analyzed in GeneSapiens.56 Normalized raw expression data were extracted from the data set, median centered and analyzed through an in-house HTML interface (REX), using R/Bioconductor-based algorithms. Associations of gene expression patterns with clinical annotations (grade, Gleason scores, invasion status) were processed with R/Bioconductor. Finally, Oncomine, a cancer microarray database and web-based data-mining platform, was used for comprehensive gene profiling of patients across different cancer types (https://www.oncomine.org/resource/login.html).44
Cell lines and monolayer cultures
Basal prostate epithelial cell line EP156T was cultured in keratinocyte serum-free medium (Life Technologies, Paisley, UK), supplemented with 50 mg/l bovine pituitary extract and 5 μg/l epidermal growth factor. PrCa cell lines were cultured in RPMI-1640 media (Life Technologies) supplemented with 10% fetal calf serum, l-glutamine (2 mm) and penicillin/streptomycin (100 μg/ml). Cells were grown in 5% CO2 at 37 °C. The EP156T cells were a gift from Varda Rotter (Weizmann Institute, Rehovot, Israel),57 LAPC4 cells were a gift from Charles Sawyer (University of California, La Jolla, CA, USA),58 whereas PC3 cells were from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells have been tested and authenticated by comparative genomic hybridization.
Western blotting
See Supplementary Information.
Miniaturized 3D cultures
Minituarized 3D cultures were prepared as described previously5, 29 (see Supplementary Information).
RNA interference
See Supplementary Information.
Live-cell imaging and confocal microscopy
Time-lapse images were acquired with Incucyte (Essen BioScience, Hertfordshire, UK), and end point imaging using confocal microscopy as described in Härmä et al.29 and Supplementary Information.
Quantitative real-time RT–PCR
See Supplementary Information.
RNA extraction and microarray hybridization
RNA extraction, labeling and hybridization were performed essentially as described in Härmä et al5 (see Supplementary Information).
Microarray data analysis
See Supplementary Information.
Inhibitor treatments
The inhibitors Y27632 and IPA3 (Tocris Bioscience, Bristol, UK) were dissolved in dimethyl sulfoxide. Treatments were performed in triplicates at 10 μm concentration and initiated at day 4 in 3D culture and finished at day 7. Multicellular structures were stained by calcein AM, imaged by spinning disk confocal microscopy, and analyzed by AMIDA, as described above.
Tumor growth assay and immunohistochemistry on the CAM
Fertilized white Leghorn chicken eggs were incubated at 37 °C under constant humidity (embryo development day 0, EDD0). No method of randomization was used. Separation of the developing CAM was induced on EDD4. On EDD8, transfected PC3 cells were suspended in PBS and Matrigel (1:1) and implanted on the CAM. On EDD11, the tumors were photographed in ovo and excised. Tumor area was measured in blind on photographs from four independent experiments, each with 3–5 eggs per shRNA construct, using ImageJ (NIH, Bethesda, MD, USA). Tumor specimens were placed in 4% paraformaldehyde, paraffin embedded and cut in 5 μm sections. Sections were deparaffinized and stained with HE. For immunohistochemistry, antigen retrieval was carried out with 10 mm sodium citrate buffer (pH 6.0). Nonspecific binding was blocked with bovine serum albumin and normal goat serum, and sections were stained with anti-MMP14 primary antibody (MAB3328; Millipore, Darmstadt, Germany) overnight at 4 °C. Sections were pretreated with 0.3% hydrogen peroxide and incubated with horse radish peroxidase-conjugated secondary antibodies (Promega, Madison, WI, USA or GE Healthcare, Buckinghamshire, England), followed by incubation with 3,3′-diaminobenzidine chromogenic substrate. Digital images were captured with Leica DM RXA light microscope (Wetzlar, Germany) and Nikon Digital Sight DS-Fi2 (Tokyo, Japan).
Statistical analysis
For RT–PCR and tumor area measurements two-tailed paired Student's t-test were used to compare groups using the Excel software (Microsoft Office). Data are presented as the mean±s.e.m. and n and the degree of significance (p-value) are specified in the figure legends. Bonferroni-corrected t-tests were used to compare groups in AMIDA analyses using the R-software environment (www.r-project.org). Box and whisker plots highlight median, data distribution, p-values, and confidence intervals. All experiments shown were replicated for at least three times. Statistical analysis for the microarray analysis of differential gene expression was performed using the limma and lumi R/Bioconductor packages.
References
Massard C, Fizazi K . Targeting continued androgen receptor signaling in prostate cancer. Clin Cancer Res 2011; 17: 3876–3883.
Epstein JI, Allsbrook WC Jr, Amin MB, Egevad LL, ISUP Grading Committee. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol 2005; 29: 1228–1242.
Friedl P, Alexander S . Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 2011; 147: 992–1009.
Olson EN, Nordheim A . Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 2010; 11: 353–365.
Härmä V, Virtanen J, Mäkelä R, Happonen A, Mpindi JP, Knuuttila M et al. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One 2010; 5: e10431.
Morin P, Wickman G, Munro J, Inman GJ, Olson MF . Differing contributions of LIMK and ROCK to TGFbeta-induced transcription, motility and invasion. Eur J Cell Biol 2011; 90: 13–25.
Härmä V, Knuuttila M, Virtanen J, Mirtti T, Kohonen P, Kovanen P et al. Lysophosphatidic acid and sphingosine-1-phosphate promote morphogenesis and block invasion of prostate cancer cells in three-dimensional organotypic models. Oncogene 2012; 31: 2075–2089.
Björk JK, Sistonen L . Regulation of the members of the mammalian heat shock factor family. FEBS J 2010; 277: 4126–4139.
Åkerfelt M, Morimoto RI, Sistonen L . Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 2010; 11: 545–555.
Dai C, Whitesell L, Rogers AB, Lindquist S . Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 2007; 130: 1005–1018.
Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 2013; 341: 1238303.
Vihervaara A, Sergelius C, Vasara J, Blom MA, Elsing AN, Roos-Mattjus P et al. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc Natl Acad Sci USA 2013; 110: E3388–E3397.
Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF . Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 2007; 26: 5086–5097.
Jin X, Moskophidis D, Mivechi NF . Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab 2011; 14: 91–103.
Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA 2011; 108: 18378–18383.
Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM et al. HSF1 Drives a Transcriptional Program Distinct from Heat Shock to Support Highly Malignant Human Cancers. Cell 2012; 150: 549–562.
Scherz-Shouval R, Santagata S, Mendillo ML, Sholl LM, Ben-Aharon I, Beck AH et al. The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell 2014; 158: 564–578.
Loison F, Debure L, Nizard P, le Goff P, Michel D, le Drean Y . Up-regulation of the clusterin gene after proteotoxic stress: implication of HSF1-HSF2 heterocomplexes. Biochem J 2006; 395: 223–231.
Östling P, Björk JK, Roos-Mattjus P, Mezger V, Sistonen L . Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem 2007; 282: 7077–7086.
Sandqvist A, Björk JK, Åkerfelt M, Chitikova Z, Grichine A, Vourc'h C et al. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol Biol Cell 2009; 20: 1340–1347.
El Fatimy R, Miozzo F, Le Mouel A, Abane R, Schwendimann L, Saberan-Djoneidi D et al. Heat shock factor 2 is a stress-responsive mediator of neuronal migration defects in models of fetal alcohol syndrome. EMBO Mol Med 2014; 6: 1043–1061.
Elsing AN, Aspelin C, Bjork JK, Bergman HA, Himanen SV, Kallio MJ et al. Expression of HSF2 decreases in mitosis to enable stress-inducible transcription and cell survival. J Cell Biol 2014; 206: 735–749.
Solimini NL, Luo J, Elledge SJ . Non-oncogene addiction and the stress phenotype of cancer cells. Cell 2007; 130: 986–988.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1.
Lee GY, Kenny PA, Lee EH, Bissell MJ . Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 2007; 4: 359–365.
Pampaloni F, Reynaud EG, Stelzer EH . The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 2007; 8: 839–845.
Härmä V, Schukov HP, Happonen A, Ahonen I, Virtanen J, Siitari H et al. Quantification of Dynamic Morphological Drug Responses in 3D Organotypic Cell Cultures by Automated Image Analysis. PLoS One 2014; 9: e96426.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4: 3.
Parri M, Chiarugi P . Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal 2010; 8: 23.
Kelly P, Casey PJ, Meigs TE . Biologic functions of the G12 subfamily of heterotrimeric g proteins: growth, migration, and metastasis. Biochemistry 2007; 46: 6677–6687.
Rasheed SA, Teo CR, Beillard EJ, Voorhoeve PM, Casey PJ . MicroRNA-182 and microRNA-200a control G-protein subunit alpha-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. J Biol Chem 2013; 288: 7986–7995.
Tcherkezian J, Lamarche-Vane N . Current knowledge of the large RhoGAP family of proteins. Biol Cell 2007; 99: 67–86.
Sinha S, Yang W . Cellular signaling for activation of Rho GTPase Cdc42. Cell Signal 2008; 20: 1927–1934.
Thiery JP, Acloque H, Huang RY, Nieto MA . Epithelial–mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Acloque H, Thiery JP, Nieto MA . The physiology and pathology of the EMT. Meeting on the epithelial–mesenchymal transition. EMBO Rep 2008; 9: 322–326.
Dagg CP, Karnofsky DA, Roddy J . Growth of transplantable human tumors in the chick embryo and hatched chick. Cancer Res 1956; 16: 589–594.
Kunzi-Rapp K, Genze F, Kufer R, Reich E, Hautmann RE, Gschwend JE . Chorioallantoic membrane assay: vascularized 3-dimensional cell culture system for human prostate cancer cells as an animal substitute model. J Urol 2001; 166: 1502–1507.
Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol 2004; 167: 769–781.
Wang X, Wilson MJ, Slaton JW, Sinha AA, Ewing SL, Pei D . Increased aggressiveness of human prostate PC-3 tumor cells expressing cell surface localized membrane type-1 matrix metalloproteinase (MT1-MMP). J Androl 2009; 30: 259–274.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004; 6: 1–6.
Ahlskog JK, Björk JK, Elsing AN, Aspelin C, Kallio M, Roos-Mattjus P et al. Anaphase-promoting complex/cyclosome participates in the acute response to protein-damaging stress. Mol Cell Biol 2010; 30: 5608–5620.
Björk JK, Sandqvist A, Elsing AN, Kotaja N, Sistonen L . miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development 2010; 137: 3177–3184.
Rossi A, Riccio A, Coccia M, Trotta E, La Frazia S, Santoro MG . The proteasome inhibitor bortezomib is a potent inducer of zinc finger AN1-type domain 2a gene expression: role of heat shock factor 1 (HSF1)-heat shock factor 2 (HSF2) heterocomplexes. J Biol Chem 2014; 289: 12705–12715.
Friedl P, Gilmour D . Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 2009; 10: 445–457.
Liotta LA, Steeg PS, Stetler-Stevenson WG . Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell 1991; 64: 327–336.
Burns-Cox N, Avery NC, Gingell JC, Bailey AJ . Changes in collagen metabolism in prostate cancer: a host response that may alter progression. J Urol 2001; 166: 1698–1701.
Shen MM, Abate-Shen C . Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010; 24: 1967–2000.
Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 1988; 80: 200–204.
Marino N, Nakayama J, Collins JW, Steeg PS . Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene. Cancer Metast Rev 2012; 31: 593–603.
Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995; 268: 884–886.
Mashimo T, Watabe M, Hirota S, Hosobe S, Miura K, Tegtmeyer PJ et al. The expression of the KAI1 gene, a tumor metastasis suppressor, is directly activated by p53. Proc Natl Acad Sci USA 1998; 95: 11307–11311.
Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol 2008; 9: R139.
Kogan I, Goldfinger N, Milyavsky M, Cohen M, Shats I, Dobler G et al. hTERT-immortalized prostate epithelial and stromal-derived cells: an authentic in vitro model for differentiation and carcinogenesis. Cancer Res 2006; 66: 3531–3540.
Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med 1997; 3: 402–408.
Hulsen T, de Vlieg J, Alkema W . BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom 2008; 9: 488.
Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res 2005; 11: 5730–5739.
Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, Reuter VE et al. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res 2006; 66: 820–827.
Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 2001; 98: 13790–13795.
Skotheim RI, Lind GE, Monni O, Nesland JM, Abeler VM, Fossa SD et al. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res 2005; 65: 5588–5598.
Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006; 9: 391–403.
Gutmann DH, Hedrick NM, Li J, Nagarajan R, Perry A, Watson MA . Comparative gene expression profile analysis of neurofibromatosis 1-associated and sporadic pilocytic astrocytomas. Cancer Res 2002; 62: 2085–2091.
Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T et al. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res 2005; 65: 3146–3154.
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 2008; 14: 518–527.
Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G . High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer 2009; 9: 152.
Adib TR, Henderson S, Perrett C, Hewitt D, Bourmpoulia D, Ledermann J et al. Predicting biomarkers for ovarian cancer using gene-expression microarrays. Br J Cancer 2004; 90: 686–692.
Durig J, Bug S, Klein-Hitpass L, Boes T, Jons T, Martin-Subero JI et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32). Leukemia 2007; 21: 2153–2163.
Haslinger C, Schweifer N, Stilgenbauer S, Dohner H, Lichter P, Kraut N et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol 2004; 22: 3937–3949.
Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 2001; 98: 1176–1181.
Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genom 2008; 1: 13.
Choi YL, Tsukasaki K, O'Neill MC, Yamada Y, Onimaru Y, Matsumoto K et al. A genomic analysis of adult T-cell leukemia. Oncogene 2007; 26: 1245–1255.
Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer 2009; 9: 11.
Acknowledgements
We thank the members of our laboratories for invaluable comments. Vidal Fey, Samu V Himanen, Heidi Lustig, Satu Orasniemi, Pauliina Toivonen, and Johannes Virtanen are acknowledged for technical assistance and other contributions. The work was supported by The Academy of Finland (FC, JKB, LS, MN, MÅ), IMI PREDECT consortium (MN, MÅ), The Sigrid Jusélius Foundation (LS), The Finnish Cancer Organizations (LS), Åbo Akademi University (JKB, LS), Åbo Akademi University Endowment Research Institute, and Tor, Joe and Pentti Borg Foundation (JKB).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Björk, J., Åkerfelt, M., Joutsen, J. et al. Heat-shock factor 2 is a suppressor of prostate cancer invasion. Oncogene 35, 1770–1784 (2016). https://doi.org/10.1038/onc.2015.241
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.241
This article is cited by
-
Gene networks and transcriptional regulators associated with liver cancer development and progression
BMC Medical Genomics (2021)
-
The proteostasis guardian HSF1 directs the transcription of its paralog and interactor HSF2 during proteasome dysfunction
Cellular and Molecular Life Sciences (2021)
-
Gambogic acid and gambogenic acid induce a thiol-dependent heat shock response and disrupt the interaction between HSP90 and HSF1 or HSF2
Cell Stress and Chaperones (2021)
-
Emerging organoid models: leaping forward in cancer research
Journal of Hematology & Oncology (2019)
-
Regulation of heat shock transcription factors and their roles in physiology and disease
Nature Reviews Molecular Cell Biology (2018)