Abstract
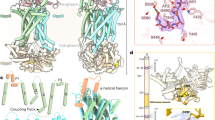
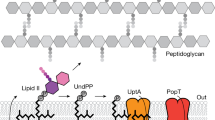
Peptidoglycan (PG) protects bacteria from osmotic lysis, and its biogenesis is a key antibiotic target. A central step in PG biosynthesis is flipping of the lipid-linked PG precursor lipid II across the cytoplasmic membrane for subsequent incorporation into PG. MurJ, part of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) transporter superfamily, was recently shown to carry out this process. However, understanding of how MurJ flips lipid II, and of how MOP transporters operate in general, remains limited due to a lack of structural information. Here we present a crystal structure of MurJ from Thermosipho africanus in an inward-facing conformation at 2.0-Å resolution. A hydrophobic groove is formed by two C-terminal transmembrane helices, which leads into a large central cavity that is mostly cationic. Our studies not only provide the first structural glimpse of MurJ but also suggest that alternating access is important for MurJ function, which may be applicable to other MOP superfamily transporters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bugg, T.D., Braddick, D., Dowson, C.G. & Roper, D.I. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 29, 167–173 (2011).
de Kruijff, B., van Dam, V. & Breukink, E. Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fatty Acids 79, 117–121 (2008).
Ruiz, N. Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 8, 21–31 (2016).
Ruiz, N. Filling holes in peptidoglycan biogenesis of Escherichia coli. Curr. Opin. Microbiol. 34, 1–6 (2016).
Mohammadi, T. et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30, 1425–1432 (2011).
Mohammadi, T. et al. Specificity of the transport of lipid II by FtsW in Escherichia coli. J. Biol. Chem. 289, 14707–14718 (2014).
Ruiz, N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. USA 105, 15553–15557 (2008).
Meeske, A.J. et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 112, 6437–6442 (2015).
Inoue, A. et al. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J. Bacteriol. 190, 7298–7301 (2008).
Sham, L.-T. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222 (2014).
Mohamed, Y.F. & Valvano, M.A. A Burkholderia cenocepacia MurJ (MviN) homolog is essential for cell wall peptidoglycan synthesis and bacterial viability. Glycobiology 24, 564–576 (2014).
Meeske, A.J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).
Hvorup, R.N. et al. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 270, 799–813 (2003).
Butler, E.K., Davis, R.M., Bari, V., Nicholson, P.A. & Ruiz, N. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J. Bacteriol. 195, 4639–4649 (2013).
Butler, E.K., Tan, W.B., Joseph, H. & Ruiz, N. Charge requirements of lipid II flippase activity in Escherichia coli. J. Bacteriol. 196, 4111–4119 (2014).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015).
He, X. et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994 (2010).
Lu, M., Radchenko, M., Symersky, J., Nie, R. & Guo, Y. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 20, 1310–1317 (2013).
Lu, M. et al. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. USA 110, 2099–2104 (2013).
Tanaka, Y. et al. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496, 247–251 (2013).
Mousa, J.J. et al. MATE transport of the E. coli–derived genotoxin colibactin. Nat. Microbiol. 1, 15009 (2016).
Fox, B.G. & Blommel, P.G. Autoinduction of protein expression. Curr. Protoc. Protein Sci. Chapter 5, Unit 5.23 (2009).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Trott, O. & Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Huber, R. et al. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 °C. Arch. Microbiol. 144, 324–333 (1986).
Boniface, A., Bouhss, A., Mengin-Lecreulx, D. & Blanot, D. The MurE synthetase from Thermotoga maritima is endowed with an unusual D-lysine adding activity. J. Biol. Chem. 281, 15680–15686 (2006).
Hsu, S.T. et al. The nisin–lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11, 963–967 (2004).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Delano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Pei, J., Kim, B.H. & Grishin, N.V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank N. Ruiz (The Ohio State University, Columbus, Ohio, USA) for sharing E. coli strains NR1154 and NR1157. We thank Z. Guan for mass spectrometry analysis of MurJ and lipid II. Data for this study were collected at beamlines NECAT 24-ID-C and 24-ID-E at the Advanced Photon Source, which are funded by grants P41GM103403 and S10 RR029205. This work was supported by Duke startup funds (S.-Y.L.).
Author information
Authors and Affiliations
Contributions
A.C.Y.K. and S.-Y.L. conceived the project. A.C.Y.K. expressed, purified, crystallized, and solved the structure of MurJTA under the guidance of S.-Y.L. E.H.M. carried out the functional complementation assays under the guidance of S.-Y.L. A.C.Y.K. and S.-Y.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Experimental phasing of MurJTA from SeMet-substituted crystals.
(a) Anomalous difference Fourier electron density peaks for selenium atoms are shown in magenta mesh contoured to 4 σ. The map was calculated from the SeMet data up to 3.5 Å resolution. (b) Initial density-modified SAD map calculated from the SeMet data up to 5.0 Å resolution without any model phases. (c) Phase-extended electron density map calculated from the SeMet data up to 3.5 Å resolution, shown here with the working model consisting of α-helical fragments. (d) The 2Fo−Fc electron density map calculated from native data up to 2.0 Å resolution with the final refined model, shown here in close-up view around the chloride site in the central cavity. The map is contoured to 1.0 σ. Water molecules are shown as gray spheres. (e) Electrostatic surface of MurJTA. Electrostatics are shown from −10 kT (red, anionic) to +10 kT (blue, cationic). The positive-inside electrostatics supports the inward-facing topology.
Supplementary Figure 2 Cl−, Zn2+, Ca2+, and Na+ sites were deduced via anomalous scattering experiments and coordination geometry.
(a) Putative ion sites were first located by anomalous scattering experiments at λ = 0.98 Å (blue mesh) and λ = 1.77 Å (red mesh). Both anomalous difference Fourier (ADF) maps were calculated to 3.5 Å resolution and contoured to 3.8 σ. (b) Cl− sites were identified by ADF density peaks from Br-soaked crystals (λ = 0.92 Å, brown mesh, calculated to 3.5 Å resolution and contoured to 4.5 σ). (c, d, e) A Zn2+ site strategically located at the start of TM7 was deduced on the basis of three factors. First, substantially higher ADF density was seen at λ = 0.98 Å (blue mesh) than at λ = 1.31 Å (green mesh) or λ = 1.77 Å (red mesh), ruling out other common divalent metals such as Co2+, Ni2+ or Fe2+. All three maps were calculated to 3.5 Å resolution and contoured to 5.5 σ. Second, the Zn2+ ion displayed restrained tetrahedral coordination geometry to two histidines and two Cl− ions, ruling out anions due to electrostatic repulsion. Third, the close coordination distance of 2.03 Å to the nitrogen of both histidines also rules out many other common metal ions. (f, g, h) Ca2+ sites were identified on the basis of: (1) ADF density peaks (λ = 1.77 Å, magenta mesh, contoured to 3.5 σ), and (2) characteristic 7-dentate pentagonal bipyramidal coordination geometry with 5 of the coordinating residues/waters arranged coplanar. The Fo−Fc omit density is shown in green mesh. Electron density for the apex water coordinating Ca2+ site 2 above the pentagonal plane is not seen. (i, j, k) Putative Na+ sites were deduced on the basis of the following: (1) lack of ADF density at λ = 1.77 Å, (2) octahedral coordination geometry with 6 coordinating groups, and (3) coordination distances of around 2.4 Å. The height of the Fo−Fc omit density (green mesh, contoured to 2.2 σ) peaks for Na+ sites 1 and 2 were respectively 4.2 and >8 σ. All omit maps were calculated from native data to 2.0 Å resolution while omitting the respective non-protein atoms. Unless stated otherwise, 2Fo−Fc maps are contoured to 1.5 σ, while Fo−Fc maps are contoured to 3.0 σ.
Supplementary Figure 3 Unmodeled electron density peaks in the portal and central cavity.
(a) The protein is sectioned on the dotted plane and viewed from the periplasm (along the direction of the arrow) to visualize the portal. The non-protein 2Fo−Fc electron density peaks are shown as magenta mesh, contoured to 0.7 σ with the protein electron density masked. Gray sticks denote monoolein molecules. (b) The protein is sectioned and viewed from the right to visualize the distal site of the central cavity. The 2Fo−Fc electron density peaks are contoured to 0.7 σ and carved 2.0 Å from the pentapeptide model. (c) Model of the pentapeptide (L-Ala-γ-D-Glu-L-Lys-D-Ala-D-Ala) built into the density in panel b, but which was not used for refinement.
Supplementary Figure 4 Western blot of total membrane fractions from NR1154 cells transformed with pEXT21 encoding either wild-type (WT) or mutant MurJTA.
MurJTA proteins carrying a C-terminal FLAG tag were expressed by the addition of 0.1 mM IPTG in the presence of arabinose. All of the mutants expressed with the exception of mutants D235A and N374A (image is representative of n = 3, technical replicates). There is no correlation between expression levels and complementation outcomes as long as the proteins are expressed: the lowest expression level was observed for the N231A mutant, which clearly complements MurJEC, while the highest expression level was observed in R24A, which clearly fails to complement. MurJTA (55 kD) migrates at an apparent molecular weight of 37 kD on a 10% SDS-PAGE gel.
Supplementary Figure 5 MurJTA is strikingly different from canonical MATE transporters, especially in TMs 1, 2, and 8, as well as in cavity electrostatics.
(a) Structural alignment between MurJTA (colored) with MATE transporters (gray) performed for individual lobes split at the start of TM7, with TMs 13 and 14 removed. Structural differences are especially pronounced at TMs 1 and 2, while TM8 of MurJTA contains a segment that deviates from the α-helical geometry observed in MATE structures. The divergence at TM7 is likely a result of the inward-facing conformation of MurJTA as compared to the outward-facing conformations of the MATE transporters. Structures of MurJTA (PDB ID:5T77), PfMATE (3VVN), NorM-VC (3MKT), NorM-NG (4HUK), DinF-BH (4LZ6), and ClbM-EC (4Z3N) were used for the alignment. (b) Surface charge differences between MurJTA and other MATE transporters. Surface electrostatics for MurJTA (PDB ID: 5T77) and MATE transporters NorM-VC (3MKT), NorM-NG (4HUK), PfMATE (3VVN), DinF-BH (4LZ6), and ClbM-EC (4Z3N) were calculated by the APBS plugin in PyMOL. The cavities of MurJTA and PfMATE are primarily cationic, in contrast to the strongly anionic cavities of NorM-VC, NorM-NG and ClbM-EC. The cavity of DinF-BH appears to have both weakly cationic and anionic patches.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Table 1 and Supplementary Note 1 (PDF 2925 kb)
Supplementary Data Set 1
Outward-facing model coordinates of MurJTA (TXT 603 kb)
Rights and permissions
About this article
Cite this article
Kuk, A., Mashalidis, E. & Lee, SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol 24, 171–176 (2017). https://doi.org/10.1038/nsmb.3346
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3346
This article is cited by
-
Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry
Nature Communications (2023)
-
Multicopy suppressor screens reveal convergent evolution of single-gene lysis proteins
Nature Chemical Biology (2023)
-
Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis
Nature Structural & Molecular Biology (2020)
-
Regulation of peptidoglycan synthesis and remodelling
Nature Reviews Microbiology (2020)
-
ECOD: identification of distant homology among multidomain and transmembrane domain proteins
BMC Molecular and Cell Biology (2019)