Key Points
-
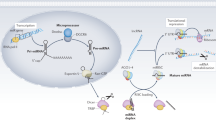
MicroRNAs (miRNAs) might have important roles in the pathogenesis and progression of IgA nephropathy
-
Abnormal expression of miR-148b in peripheral blood mononuclear cells might account for the aberrant glycosylation of IgA1 observed in patients with IgA nephropathy
-
miR-29c attenuates renal interstitial fibrosis and is probably important in the progression of IgA nephropathy
-
Urinary miRNA levels could potentially serve as biomarkers for diagnosing and monitoring IgA nephropathy
-
The potential application of urinary miRNAs in monitoring patients with IgA nephropathy is still in the early stages of development, with available published evidence limited to small-scale studies
Abstract
IgA nephropathy is globally the most common primary glomerulonephritis, but the pathogenesis of this condition is still only partially understood. MicroRNAs (miRNAs) are short, noncoding RNA molecules that regulate gene expression. Genome-wide analysis of renal miRNA expression has identified a number of novel miRNAs related to immunological and pathological changes. Specifically, overexpression of miR-148b might explain the aberrant glycosylation of IgA1, which has a central pathogenetic role in the early phase of IgA nephropathy. By contrast, miR-29c is an antifibrotic miRNA that is probably important in the late stages of disease progression. In addition, urinary levels of several miRNAs are significantly changed in patients with IgA nephropathy compared with healthy individuals; some alterations seem to be disease-specific, whereas others are apparently damage-related. As miRNAs in urinary sediment are relatively stable and easily quantified, they have the potential to be used as biomarkers for the diagnosis and monitoring of disease. However, to date, limited data are available on the role of miRNAs in the pathogenesis of IgA nephropathy and their potential application as biomarkers. Consequently, further studies are urgently needed to address this shortfall. Here, we review the available literature on miRNAs in relation to IgA nephropathy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Wyatt, R. J. & Julian, B. A. IgA nephropathy. N. Engl. J. Med. 368, 2402–2414 (2013).
Li, P. K., Ho, K. K., Szeto, C. C., Yu, L. & Lai, F. M. Prognostic indicators of IgA nephropathy in the Chinese—clinical and pathological perspectives. Nephrol. Dial. Transplant. 17, 64–69 (2002).
Le, W. et al. Long-term renal survival and related risk factors in patients with IgA nephropathy: results from a cohort of 1155 cases in a Chinese adult population. Nephrol. Dial. Transplant. 27, 1479–1485 (2012).
Zuo, L. & Wang, M. Current burden and probable increasing incidence of ESRD in China. Clin. Nephrol. 74 (Suppl. 1), S20–S22 (2010).
Schena, F. P. & Coppo, R. in Oxford Textbook of Clinical Nephrology 3rd edn (eds Davison, A. M. et al.) 469–501 (Oxford University Press, 2005).
Ho, Y.-W. et al. Hong Kong Renal Registry Report 2010. Hong Kong J. Nephrol. 12, 81–98 (2010).
Knoop, T. et al. Mortality in patients with IgA nephropathy. Am. J. Kidney Dis. 62, 883–890 (2013).
Novak, J., Renfrow, M. B., Gharavi, A. G. & Julian, B. A. Pathogenesis of immunoglobulin A nephropathy. Curr. Opin. Nephrol. Hypertens. 22, 287–294 (2013).
Roos, A. & van Kooten, C. Underglycosylation of IgA in IgA nephropathy: more than a diagnostic marker? Kidney Int. 71, 1089–1091 (2007).
Boyd, J. K., Cheung, C. K., Molyneux, K., Feehally, J. & Barratt, J. An update on the pathogenesis and treatment of IgA nephropathy. Kidney Int. 81, 833–843 (2012).
Moldoveanu, Z. et al. Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 71, 1148–1154 (2007).
Smith, A. C., Molyneux, K., Feehally, J. & Barratt, J. O-glycosylation of serum IgA1 antibodies against mucosal and systemic antigens in IgA nephropathy. J. Am. Soc. Nephrol. 17, 3520–3528 (2006).
Li, G. S., Zhang, H., Lv, J. C., Shen, Y. & Wang, H. Y. Variants of C1GALT1 gene are associated with the genetic susceptibility to IgA nephropathy. Kidney Int. 71, 448–453 (2007).
Serino, G., Sallustio, F., Cox, S. N., Pesce, F. & Schena, F. P. Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J. Am. Soc. Nephrol. 23, 814–824 (2012).
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349 (2004).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116, 281–297 (2004).
Morita, K. & Han, M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 25, 5794–5804 (2006).
Chan, S. P., Ramaswamy, G., Choi, E. Y. & Slack, F. J. Identification of specific let-7 microRNA binding complexes in Caenorhabditis elegans. RNA 14, 2104–2114 (2008).
Bernstein, E. et al. Dicer is essential for mouse development. Nat. Genet. 35, 215–217 (2003).
Bartels, C. L. & Tsongalis, G. J. MicroRNAs, novel biomarkers for human cancer. Clin. Chem. 55, 623–631 (2009).
Xiao, C. & Rajewsky, K. MicroRNA control in the immune system, basic principles. Cell 136, 26–36 (2009).
Tian, Z., Greene, A. S., Pietrusz, J. L., Matus, I. R. & Liang, M. MicroRNA–target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 18, 404–411 (2008).
Sun, Y. et al. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 32, e188 (2004).
Gregory, P. A., Bracken, C. P., Bert, A. G. & Goodall, G. J. MicroRNAs as regulators of epithelial–mesenchymal transition. Cell Cycle 7, 3112–3118 (2008).
Neilson, E. G. Mechanisms of disease: fibroblasts—a new look at an old problem. Nat. Clin. Pract. Nephrol. 2, 101–108 (2006).
Burk, U. et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 (2008).
Park, S.-M., Gaur, A. B., Lengyel, E. & Peter, M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 (2008).
Korpal, M., Lee, E. S., Hu, G. & Kang, Y. The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 (2008).
Bracken, C. P. et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res. 68, 7846–7854 (2008).
Gregory, P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 (2008).
Lorenzen, J. M., Haller, H. & Thum, T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat. Rev. Nephrol. 7, 286–294 (2011).
Chandrasekaran, K. et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 81, 617–627 (2012).
Tan, K. et al. Genome-wide analysis of microRNAs expression profiling in patients with primary IgA nephropathy. Genome 56, 161–169 (2013).
Tanzer, A. & Stadler, P. F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 339, 327–335 (2004).
Cloonan, N. et al. The miR-17-5p microRNA is a key regulator of the G1/S. phase cell cycle transition. Genome Biol. 9, R127 (2008).
Serva, A. et al. miR-17-5p regulates endocytic trafficking through targeting TBC1D2/Armus. PLoS ONE 7, e52555 (2012).
Dong, Y. et al. Tumor suppressor functions of miR-133a in colorectal cancer. Mol. Cancer Res. 11, 1051–1060 (2013).
Liu, W. et al. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 9, e1003626 (2013).
Wang, J. et al. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 4, e699 (2013).
Silva, F. G., Chander, P., Pirani, C. L. & Hardy, M. A. Disappearance of glomerular mesangial IgA deposits after renal allograft transplantation. Transplantation 33, 241–246 (1982).
Chen, Y, Song, Y. X. & Wang, Z. N. The microRNA-148/152 family, multi-faceted players. Mol. Cancer 12, 43 (2013).
Liu, X. et al. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J. Immunol. 185, 7244–7251 (2010).
Cimino, D. et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 27, 1223–1235 (2013).
Roth, C. et al. Low levels of cell-free circulating miR-361-3p and miR-625* as blood-based markers for discriminating malignant from benign lung tumors. PLoS ONE 7, e38248 (2012).
Cao, J. et al. DNA methylation-mediated repression of miR-886-3p predicts poor outcome of human small cell lung cancer. Cancer Res. 73, 3326–3335 (2013).
Floege, J. The pathogenesis of IgA nephropathy, what is new and how does it change therapeutic approaches? Am. J. Kidney Dis. 58, 992–1004 (2011).
Loeffler, I. & Wolf, G. Transforming growth factor-β and the progression of renal disease. Nephrol. Dial. Transplant. 29 (Suppl. 1), i37–i45 (2014).
Liu, Y. et al. Renal medullary microRNAs in Dahl salt-sensitive rats, miR-29b regulates several collagens and related genes. Hypertension 55, 974–982 (2010).
Qin, W. et al. TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J. Am. Soc. Nephrol. 22, 1462–1474 (2011).
Fang, Y. et al. miR-29c is downregulated in renal interstitial fibrosis in humans and rats and restored by HIF-α activation. Am. J. Physiol. Renal Physiol. 304, F1274–F1282 (2013).
Wang, G. et al. Intra-renal expression of microRNAs in patients with IgA nephropathy. Lab. Invest. 90, 98–103 (2010).
Wang, G. et al. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis. Markers 30, 171–179 (2011).
Puhr, M. et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am. J. Pathol. 181, 2188–2201 (2012).
Muratsu-Ikeda, S. et al. Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PLoS ONE 7, e41462 (2012).
Yang, L. et al. miR-146a controls the resolution of T cell responses in mice. J. Exp. Med. 209, 1655–1670 (2012).
Ichii, O. et al. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int. 81, 280–292 (2012).
Vigorito, E., Kohlhaas, S., Lu, D. & Leyland, R. miR-155, an ancient regulator of the immune system. Immunol. Rev. 253, 146–157 (2013).
Chau, B. N. et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 4, 121ra128 (2012).
Chung, A. C., Huang, X. R., Meng, X. & Lan, H. Y. miR-192 mediates TGF-β/Smad3-driven renal fibrosis. J. Am. Soc. Nephrol. 21, 1317–1325 (2010).
Krupa, A., Jenkins, R., Luo, D. D., Lewis, A., Phillips, A. & Fraser, D. Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J. Am. Soc. Nephrol. 21, 438–447 (2010).
Du, B. et al. High glucose down-regulates miR-29a to increase collagen IV production in HK-2 cells. FEBS Lett. 584, 811–816 (2010).
Wang, Q. et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 22, 4126–4135 (2008).
Fleissner, F. et al. Short communication, asymmetric dimethylarginine impairs angiogenic progenitor cell function in patients with coronary artery disease through a microRNA-21-dependent mechanism. Circ. Res. 107, 138–143 (2010).
Hanke, M. et al. A robust methodology to study urine microRNA as tumor marker, microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 28, 655–661 (2010).
Cortez, M. A. & Calin, G. A. MicroRNA identification in plasma and serum, a new tool to diagnose and monitor diseases. Expert Opin. Biol. Ther. 9, 703–711 (2009).
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA 105, 10513–10518 (2008).
Gilad, S. et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 3, e3148 (2008).
Lorenzen, J. M. et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am. J. Transplant. 11, 2221–2227 (2011).
Wang, G. et al. Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus 20, 493–500 (2011).
Wang, G. & Szeto, C. C. Methods of microRNA quantification in urinary sediment. Methods Mol. Biol. 1024, 211–220 (2013).
Turchinovich, A., Weiz, L., Langheinz, A. & Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 (2011).
Wang, G. et al. Serum and urinary cell free miR-146a and miR-155 in patients with systemic lupus erythematosus. J. Rheumatol. 37, 2516–2522 (2010).
Wang, G. et al. Urinary miR-21, miR-29, and miR-93, novel biomarkers of fibrosis. Am. J. Nephrol. 36, 412–418 (2012).
Szeto, C. C. et al. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Dis. Markers 33, 137–144 (2012).
Wang, G. et al. Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis. Markers 28, 79–86 (2010).
Acknowledgements
C.-C.S. is supported in part by the Chinese University of Hong Kong research account 6901031.
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, discussed its content, wrote the article, reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
P.K.-T.L. has received speaker honoraria from Astellas and sits on the Trial Advisory Committee of Baxter Healthcare. C.-C.S. has received research grants from Baxter Healthcare.
Rights and permissions
About this article
Cite this article
Szeto, CC., Li, PT. MicroRNAs in IgA nephropathy. Nat Rev Nephrol 10, 249–256 (2014). https://doi.org/10.1038/nrneph.2014.50
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneph.2014.50
This article is cited by
-
Urinary exosomal miRNA signature of IgA nephropathy: a case–control study
Scientific Reports (2023)
-
Blood TGF-β1 and miRNA-21-5p levels predict renal fibrosis and outcome in IgA nephropathy
International Urology and Nephrology (2023)
-
ICAM-1 related long noncoding RNA is associated with progression of IgA nephropathy and fibrotic changes in proximal tubular cells
Scientific Reports (2022)
-
Recent findings on the role of microRNAs in genetic kidney diseases
Molecular Biology Reports (2022)
-
Urinary miRNA profile for the diagnosis of IgA nephropathy
BMC Nephrology (2019)