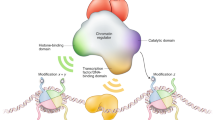
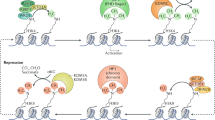
Key Points
-
The identification of the Lys-specific demethylase (LSD) and the Jumonji C (JMJC) protein families that catalyse the demethylation of methylated Lys residues has led to a new paradigm for how histone methylation is regulated.
-
The histone demethylases are involved in regulating cellular processes such as chromatin structure and transcription. They are important for normal embryonic development and are involved in diseases such as cancer.
-
Both histones and non-histone proteins are targets for the histone demethylases. Examples of non-histone substrates include p53, DNA methyltransferase 1 (DNMT1) and E2F1.
-
Histone demethylase activity is regulated by expression levels, by targeting to specific sites in the genome and by post-translational modifications.
-
Histone demethylases bind to many target genes, where they enable transcriptional competence by ensuring a specific chromatin state.
Abstract
Histone modifications are thought to regulate chromatin structure, transcription and other nuclear processes. Histone methylation was originally believed to be an irreversible modification that could only be removed by histone eviction or by dilution during DNA replication. However, the isolation of two families of enzymes that can demethylate histones has changed this notion. The biochemical activities of these histone demethylases towards specific Lys residues on histones, and in some cases non-histone substrates, have highlighted their importance in developmental control, cell-fate decisions and disease. Their ability to be regulated through protein-targeting complexes and post-translational modifications is also beginning to shed light on how they provide dynamic control during transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Kuo, M. H. & Allis, C. D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20, 615–626 (1998).
Khan, S. N. & Khan, A. U. Role of histone acetylation in cell physiology and diseases: an update. Clin. Chim. Acta 411, 1401–1411 (2010).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). The first paper to describe histone demethylase activity.
Barth, T. K. & Imhof, A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 35, 618–626 (2010).
Albert, M. & Helin, K. Histone methyltransferases in cancer. Semin. Cell Dev. Biol. 21, 209–220 (2010).
Pedersen, M. T. & Helin, K. Histone demethylases in development and disease. Trends Cell Biol. 20, 662–671 (2010).
Klose, R. J., Kallin, E. M. & Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev. Genet. 7, 715–727 (2006).
Chen, X., Hu, Y. & Zhou, D.-X. Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809, 421–426 (2011).
Schotta, G. et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22, 2048–2061 (2008).
Jung, H. R., Pasini, D., Helin, K. & Jensen, O. N. Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol. Cell. Proteomics 9, 838–850 (2010).
Garcia, B. A. et al. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282, 7641–7655 (2007).
Hake, S. B. et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281, 559–568 (2006).
Waldmann, T. et al. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11 (2011).
Gardner, K. E., Zhou, L., Parra, M. A., Chen, X. & Strahl, B. D. Identification of lysine 37 of histone H2B as a novel site of methylation. PLoS ONE 6, e16244 (2011).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Huang, J. et al. p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007).
Wang, J. et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nature Genet. 41, 125–129 (2009). These authors identified a link between histone demethylation by LSD1 and DNA methylation.
Kontaki, H. & Talianidis, I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell 39, 152–160 (2010).
Karytinos, A. et al. A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 284, 17775–17782 (2009).
Ciccone, D. N. et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418 (2009).
Lee, M. G. et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450 (2007).
Cho, Y.-W. et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406 (2007).
Stavropoulos, P., Blobel, G. & Hoelz, A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nature Struct. Mol. Biol. 13, 626–632 (2006).
Issaeva, I. et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 27, 1889–1903 (2007).
Mohan, M. et al. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 31, 4310–4318 (2011).
Lee, M. G., Wynder, C., Cooch, N. & Shiekhattar, R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 (2005).
Tsukada, Y.-I. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006). The first published paper describing a member of the JMJC domain family as a histone demethylase.
Klose, R. J. et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 (2006).
Cloos, P. A. et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442, 307–311 (2006).
Fodor, B. D. et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20, 1557–1562 (2006).
Whetstine, J. R. et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 (2006). References 30–33 were the first papers to describe trimethyl demethylases.
Christensen, J. et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128, 1063–1076 (2007).
Agger, K. et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734 (2007).
Lan, F. et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694 (2007).
De Santa, F. et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 (2007).
Eissenberg, J. C. et al. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nature Struct. Mol. Biol. 14, 344–346 (2007).
Seward, D. J. et al. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nature Struct. Mol. Biol. 14, 240–242 (2007).
Secombe, J., Li, L., Carlos, L. & Eisenman, R. N. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21, 537–551 (2007).
Liang, G., Klose, R. J., Gardner, K. E. & Zhang, Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nature Struct. Mol. Biol. 14, 243–245 (2007).
Huarte, M. et al. The fission yeast Jmj2 reverses histone H3 Lysine 4 trimethylation. J. Biol. Chem. 282, 21662–21670 (2007).
Kim, T. & Buratowski, S. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J. Biol. Chem. 282, 20827–20835 (2007).
Tahiliani, M. et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447, 601–605 (2007).
Frescas, D., Guardavaccaro, D., Bassermann, F., Koyama-Nasu, R. & Pagano, M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450, 309–313 (2007).
Yamane, K. et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 (2006).
Iwase, S. et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088 (2007).
Klose, R. J. et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128, 889–900 (2007).
Lee, M. G., Norman, J., Shilatifard, A. & Shiekhattar, R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128, 877–887 (2007).
Liu, W. et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466, 508–512 (2010).
Duncan, E. M. & Allis, C. D. Errors in erasure: links between histone lysine methylation removal and disease. Prog. Drug Res. 67, 69–90 (2010).
Yoshimi, A. & Kurokawa, M. Key roles of histone methyltransferase and demethylase in leukemogenesis. J. Cell Biochem. 112, 415–424 (2011).
Di Stefano, L., Ji, J.-Y., Moon, N.-S., Herr, A. & Dyson, N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr. Biol. 17, 808–812 (2007).
Eliazer, S., Shalaby, N. A. & Buszczak, M. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc. Natl Acad. Sci. USA 108, 7064–7069 (2011).
Katz, D. J., Edwards, T. M., Reinke, V. & Kelly, W. G. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137, 308–320 (2009).
Gildea, J. J., Lopez, R. & Shearn, A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156, 645–663 (2000).
Greer, E. L. et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479, 365–371 (2011).
Wang, J. et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887 (2007).
Okada, Y., Scott, G., Ray, M. K., Mishina, Y. & Zhang, Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450, 119–123 (2007).
Inagaki, T. et al. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 14, 991–1001 (2009).
Tateishi, K., Okada, Y., Kallin, E. M. & Zhang, Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458, 757–761 (2009).
Liu, Z. et al. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J. Biol. Chem. 285, 2758–2770 (2010).
Zhang, Q.-J. et al. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J. Clin. Invest. 121, 2447–2456 (2011).
Iwamori, N., Zhao, M., Meistrich, M. L. & Matzuk, M. M. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol. Reprod. 84, 1225–1234 (2011).
Catchpole, S. et al. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int. J. Oncol. 38, 1267–1277 (2011).
Schmitz, S. U. et al. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 30, 4586–4600 (2011).
Takeuchi, T. et al. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 9, 1211–1222 (1995). A historically interesting paper in which the founding member of the JMJ protein family was characterized as being important for normal development.
Motoyama, J., Kitajima, K., Kojima, M., Kondo, S. & Takeuchi, T. Organogenesis of the liver, thymus and spleen is affected in jumonji mutant mice. Mech. Dev. 66, 27–37 (1997).
Takeuchi, T., Kojima, M., Nakajima, K. & Kondo, S. Jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech. Dev. 86, 29–38 (1999).
Baker, R. K. et al. In vitro preselection of gene-trapped embryonic stem cell clones for characterizing novel developmentally regulated genes in the mouse. Dev. Biol. 185, 201–214 (1997).
Böse, J. et al. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 3, 15 (2004).
Kunisaki, Y. et al. Clearance receptor controls death and differentiation. Blood 103, 3250–3250 (2004).
Li, M. O. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 302, 1560–1563 (2003).
Fukuda, T., Tokunaga, A., Sakamoto, R. & Yoshida, N. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol. Cell. Neurosci. 46, 614–624 (2011).
Ishimura, A. et al. Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development 139, 749–759 (2012).
Satoh, T. et al. The Jmjd3–Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nature Immunol. 11, 936–944 (2010).
Cox, B. J. et al. Phenotypic annotation of the mouse X chromosome. Genome Res. 20, 1154–1164 (2010).
Lee, S., Lee, J. W. & Lee, S.-K. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell 22, 25–37 (2012).
Okada, Y., Tateishi, K. & Zhang, Y. Histone demethylase JHDM2A is involved in male infertility and obesity. J. Androl. 31, 75–78 (2010).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Kahl, P. et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 66, 11341–11347 (2006).
Schulte, J. H. et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 69, 2065–2071 (2009).
Lim, S. et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31, 512–520 (2010).
Wang, Y. et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672 (2009).
Abidi, F. E. et al. Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. J. Med. Genet. 45, 787–793 (2008).
Jensen, L. R. et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236 (2005).
Rujirabanjerd, S. et al. Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype-phenotype correlation. Eur. J. Hum. Genet. 18, 330–335 (2010).
Santos, C. et al. A novel mutation in JARID1C gene associated with mental retardation. Eur. J. Hum. Genet. 14, 583–586 (2006).
Adegbola, A., Gao, H., Sommer, S. & Browning, M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD). Am. J. Med. Genet. A 146, 505–511 (2008).
Santos-Rebouças, C. B. et al. A novel nonsense mutation in KDM5C/JARID1C gene causing intellectual disability, short stature and speech delay. Neurosci. Lett. 498, 67–71 (2011).
Tzschach, A. et al. Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum. Mutat. 27, 389 (2006).
Jensen, L. R. et al. A distinctive gene expression fingerprint in mentally retarded male patients reflects disease-causing defects in the histone demethylase KDM5C. Pathogenetics 3, 2 (2010).
Yang, M. et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 23, 377–387 (2006).
Yang, M. et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nature Struct. Mol. Biol. 14, 535–539 (2007).
Forneris, F., Binda, C., Adamo, A., Battaglioli, E. & Mattevi, A. Structural basis of LSD1–CoREST selectivity in histone H3 recognition. J. Biol. Chem. 282, 20070–20074 (2007).
Chen, Z. et al. Structural insights into histone demethylation by JMJD2 family members. Cell 125, 691–702 (2006).
Ng, S. S. et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448, 87–91 (2007).
Couture, J.-F., Collazo, E., Ortiz-Tello, P. A., Brunzelle, J. S. & Trievel, R. C. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nature Struct. Mol. Biol. 14, 689–695 (2007).
Chen, Z. et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl Acad. Sci. USA 104, 10818–10823 (2007).
Horton, J. R. et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nature Struct. Mol. Biol. 17, 38–43 (2009). An elegant study that used crystallography on closely related demethylases (PHF8 and KIAA1718) to determine how substrate specificity is established.
Kleine-Kohlbrecher, D. et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38, 165–178 (2010).
Loenarz, C. et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum. Mol. Genet. 19, 217–222 (2010).
Yu, L. et al. Structural insights into a novel histone demethylase PHF8. Cell Res. 20, 166–173 (2010).
Huang, C. et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 20, 154–165 (2010).
Tsukada, Y.-I., Ishitani, T. & Nakayama, K. I. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 24, 432–437 (2010).
Young, N. L. et al. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics 8, 2266–2284 (2009).
Yang, Y. et al. Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res. 20, 886–898 (2010).
Lin, H. et al. Coordinated regulation of active and repressive histone methylations by a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res. 20, 899–907 (2010).
Wissmann, M. et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nature Cell Biol. 9, 347–353 (2007).
Shin, S. & Janknecht, R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem. Biophys. Res. Commun. 359, 742–746 (2007).
Xiang, Y. et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl Acad. Sci. USA 104, 19226–19231 (2007).
Agger, K. et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A–ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 23, 1171–1176 (2009).
Barradas, M. et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 23, 1177–1182 (2009).
Shi, Y.-J. et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005).
Adamo, A. et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nature Cell Biol. 13, 652–659 (2011).
Gordon, M. et al. Genome-wide dynamics of SAPHIRE, an essential complex for gene activation and chromatin boundaries. Mol. Cell. Biol. 27, 4058–4069 (2007).
Fang, R. et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell 39, 222–233 (2010).
Blackledge, N. P. et al. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell 38, 179–190 (2010). Showed that CpG islands can be directly involved in the recruitment of histone demethylases and thereby help to delineate the architecture of chromatin at CpG islands.
Chen, H., Kluz, T., Zhang, R. & Costa, M. Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis 31, 2136–2144 (2010).
Roesch, A. et al. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int. J. Cancer 122, 1047–1057 (2008).
Xie, L. et al. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 30, 1473–1484 (2011).
De Santa, F. et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28, 3341–3352 (2009).
Fortschegger, K. et al. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol. Cell. Biol. 30, 3286–3298 (2010).
Qi, H. H. et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507 (2010).
Whyte, W. A. et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012).
Lin, C.-H. et al. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol. Cell 32, 696–706 (2008).
He, J., Nguyen, A. T. & Zhang, Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 117, 3869–3880 (2011).
Kwon, D.-W. & Ahn, S. H. Role of yeast JmjC-domain containing histone demethylases in actively transcribed regions. Biochem. Biophys. Res. Commun. 410, 614–619 (2011).
Fang, J., Hogan, G. J., Liang, G., Lieb, J. D. & Zhang, Y. The Saccharomyces cerevisiae histone demethylase Jhd1 fine-tunes the distribution of H3K36me2. Mol. Cell. Biol. 27, 5055–5065 (2007).
Tu, S. et al. Identification of histone demethylases in Saccharomyces cerevisiae. J. Biol. Chem. 282, 14262–14271 (2007).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Solá, S. et al. p53 interaction with JMJD3 results in its nuclear distribution during mouse neural stem cell differentiation. PLoS ONE 6, e18421 (2011).
Xie, Q. et al. Methylation-mediated regulation of E2F1 in DNA damage-induced cell death. J. Recept. Signal Transduct. Res. 31, 139–146 (2011).
Lu, T. et al. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl Acad. Sci. USA 107, 46–51 (2010).
Lu, T. et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NF-κB. Proc. Natl Acad. Sci. USA 106, 16339–16344 (2009).
Landeira, D. & Fisher, A. G. Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol. 21, 74–80 (2011).
Peng, J. C. et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139, 1290–1302 (2009).
Shen, X. et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139, 1303–1314 (2009).
Pasini, D. et al. JARID2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 (2010). References 137, 138 and 139 describe the role of JARID2 in recruiting PRC2 in ES cells.
Chang, B., Chen, Y., Zhao, Y. & Bruick, R. K. JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007).
Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009).
Hahn, P. et al. Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation. PLoS ONE 5, e13769 (2010).
Miller, S. A., Mohn, S. E. & Weinmann, A. S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605 (2010).
DiTacchio, L. et al. Histone lysine demethylase JARID1a activates CLOCK–BMAL1 and influences the circadian clock. Science 333, 1881–1885 (2011).
Huang, Y., Fang, J., Bedford, M. T., Zhang, Y. & Xu, R.-M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312, 748–751 (2006).
Lee, J., Thompson, J. R., Botuyan, M. V. & Mer, G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nature Struct. Mol. Biol. 15, 109–111 (2008).
Ozboyaci, M., Gursoy, A., Erman, B. & Keskin, O. Molecular recognition of H3/H4 histone tails by the tudor domains of JMJD2A: a comparative molecular dynamics simulations study. PLoS ONE 6, e14765 (2011).
Yap, K. L. & Zhou, M. M. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit. Rev. Biochem. Mol. Biol. 45, 488–505 (2010).
Patsialou, A., Wilsker, D. & Moran, E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33, 66–80 (2005).
Tu, S. et al. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nature Struct. Mol. Biol. 15, 419–421 (2008).
Ruthenburg, A. J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Saleque, S., Kim, J., Rooke, H. M. & Orkin, S. H. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 27, 562–572 (2007).
Mulligan, P. et al. A SIRT1–LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell 42, 689–699 (2011).
Shi, L. et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl Acad. Sci. USA 108, 7541–7546 (2011).
Black, J. C. et al. Conserved antagonism between JMJD2A/KDM4A and HP1-γ during cell cycle progression. Mol. Cell 40, 736–748 (2010).
Di Stefano, L. et al. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 25, 17–28 (2011).
Rudolph, T. et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol. Cell 26, 103–115 (2007).
Baba, A. et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2–ARID5B. Nature Cell Biol. 13, 668–675 (2011). The authors show that the activity of the histone demethylase PHF2 depends on its phosphorylation status and therefore is subject to regulation by post-translational modification.
Krishnakumar, R. & Kraus, W. L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 (2010).
Van Rechem, C. et al. The SKP1–Cul1–F-box and leucine-rich repeat protein 4 (SCF–FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/jumonji domain-containing 2A (JMJD2A) protein. J. Biol. Chem. 286, 30462–30470 (2011).
Tan, M.-K. M., Lim, H.-J. & Harper, J. W. SCFFBXO22 regulates histone H3 Lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol. Cell. Biol. 31, 3687–3699 (2011).
D'Amours, D., Desnoyers, S., D'Silva, I. & Poirier, G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 (1999).
Rouleau, M., Patel, A., Hendzel, M. J., Kaufmann, S. H. & Poirier, G. G. PARP inhibition: PARP1 and beyond. Nature Rev. Cancer 10, 293–301 (2010).
Ji, Y. & Tulin, A. V. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 20, 512–518 (2010).
Krishnakumar, R. & Kraus, W. L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 (2010).
Henikoff, S. & Shilatifard, A. Histone modification: cause or cog? Trends Genet. 27, 389–396 (2011).
Lee, B. M. & Mahadevan, L. C. Stability of histone modifications across mammalian genomes: implications for “epigenetic” marking. J. Cell. Biochem. 108, 22–34 (2009).
Hayami, S. et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer 128, 574–586 (2011).
Kauffman, E. C. et al. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol. Carcinog. 50, 931–944 (2011).
Heidenblad, M. et al. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med. Genomics 1, 3 (2008).
Köninger, J. et al. Phosphatidylserine receptor in chronic pancreatitis: evidence for a macrophage independent role. Ann. Surg. 241, 144–151 (2005).
Kottakis, F. et al. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101–EZH2 pathway. Mol. Cell 43, 285–298 (2011).
Sinha, S. et al. Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at chromosomal 9q22.3 region: pathological significance in early- and late-onset breast carcinoma. Mol. Cancer 7, 84 (2008).
Ghosh, A. et al. Association of FANCC and PTCH1 with the development of early dysplastic lesions of the head and neck. Ann. Surg. Oncol. 23 Aug 2011 (doi:10.1245/s10434-011-1991-x).
Laumonnier, F. et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 42, 780–786 (2005).
Abidi, F. E., Miano, M. G., Murray, J. C. & Schwartz, C. E. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin. Genet. 72, 19–22 (2007).
Koivisto, A. M. et al. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin. Genet. 72, 145–149 (2007).
Uemura, M. et al. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: in vivo identification from hypoxic tumor cells. Clin. Cancer Res. 16, 4636–4646 (2010).
Qi, J. et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 18, 23–38 (2010).
Guo, X. et al. The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma 58, 153–157 (2011).
Yamada, D. et al. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection. Ann. Surg. Oncol. 24 May 2011 (doi:10.1245/s10434-011-1797-x).
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 41, 521–523 (2009).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Gui, Y. et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature Genet. 43, 875–878 (2011).
Jankowska, A. M. et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 118, 3932–3941 (2011).
Patani, N., Jiang, W. G., Newbold, R. F. & Mokbel, K. Histone-modifier gene expression profiles are associated with pathological and clinical outcomes in human breast cancer. Anticancer Res. 31, 4115–4125 (2011).
Lederer, D. et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with kabuki syndrome. Am. J. Hum. Genet. 90, 119–124 (2012).
Ciavatta, D. J. et al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J. Clin. Invest. 120, 3209–3219 (2010).
Anderton, J. A. et al. The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus and over-expressed in Hodgkin's Lymphoma. Oncogene 30, 2037–2043 (2011).
Pryor, J. G., Brown-Kipphut, B. A., Iqbal, A. & Scott, G. A. Microarray comparative genomic hybridization detection of copy number changes in desmoplastic melanoma and malignant peripheral nerve sheath tumor. Am. J. Dermatopathol. 33, 780–785 (2011).
Yang, Z. Q. et al. Identification of a novel gene, GASC1, within an amplicon at 9p23–24 frequently detected in esophageal cancer cell lines. Cancer Res. 60, 4735–4739 (2000).
Liu, G. et al. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 28, 4491–4500 (2009).
Ehrbrecht, A. et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J. Pathol. 208, 554–563 (2006).
Vinatzer, U. et al. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin. Cancer Res. 14, 6426–6431 (2008).
Vogt, T. et al. Deficiency of a novel retinoblastoma binding protein 2-homolog is a consistent feature of sporadic human melanoma skin cancer. Lab. Invest. 79, 1615–1627 (1999).
van Zutven, L. J. et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer 45, 437–446 (2006).
Pointon, J. J. et al. The histone demethylase JARID1A is associated with susceptibility to ankylosing spondylitis. Genes Immun. 12, 395–398 (2011).
Hayami, S. et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol. Cancer 9, 59 (2010).
Lu, P. J. et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274, 15633–15645 (1999).
Barrett, A. et al. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen? Int. J. Cancer 101, 581–588 (2002).
Perinchery, G. et al. Deletion of Y-chromosome specific genes in human prostate cancer. J. Urol. 163, 1339–1342 (2000).
Lee, Y. et al. Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res. 86, 932–938 (2000).
Mysliwiec, M. R. et al. Generation of a conditional null allele of jumonji. Genesis 44, 407–411 (2006).
Scapoli, L. et al. Expression and association data strongly support JARID2 involvement in nonsyndromic cleft lip with or without cleft palate. Hum. Mutat. 31, 794–800 (2010).
Volcik, K. A. et al. Evaluation of the jumonji gene and risk for spina bifida and congenital heart defects. Am. J. Med. Genet. 126A, 215–217 (2004).
Suzuki, C. et al. Identification of Myc-associated protein with JmjC domain as a novel therapeutic target oncogene for lung cancer. Mol. Cancer Ther. 6, 542–551 (2007).
Acknowledgements
The authors thank M. T. Pedersen and I. Comet for providing critical and constructive comments on the manuscript and members of the Helin laboratory for discussions. S.M.K is supported by a postdoctoral fellowship from the Netherlands Organization for Scientific Research (NWO). Work in the Helin laboratory is supported by the Danish National Research Foundation, the Danish Cancer Society, the Novo Nordisk Foundation, Denmark, the Danish Medical Research Council, the Lundbeck Foundation, Denmark, the European Union and the Excellence Programme of the University of Copenhagen.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Gastrulate
-
Gastulation is a phase in early embryonic development of animals during which the embryo is reorganized into the three germ layers — ectoderm, endoderm and mesoderm.
- Imprinting
-
A genomic mechanism for the exclusive expression of certain genes from either the maternal or the paternal allele.
- Plant homeodomain
-
(PHD). A zinc-finger-containing protein domain that is involved in mediating protein–protein interactions. It can recognize differently modified nucleosomes and is often found in chromatin regulators.
- Chromatin immunoprecipitation
-
(ChIP). A method that allows isolation of DNA sequences that are bound to a protein of interest in vivo using specific antibodies to the protein.
- Ubiquitin ligase complex
-
A protein complex that is responsible for the addition of ubiquitin to Lys residues within target proteins. The covalent ubiquitin modification influences the fate and function of its targets.
- Proteasome
-
A large multisubunit protease complex (26S) that selectively degrades polyubiquitylated proteins. It contains a 20S particle that carries the catalytic activity and two regulatory 19S particles.
Rights and permissions
About this article
Cite this article
Kooistra, S., Helin, K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13, 297–311 (2012). https://doi.org/10.1038/nrm3327
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3327
This article is cited by
-
Deubiquitinase USP7 stabilizes KDM5B and promotes tumor progression and cisplatin resistance in nasopharyngeal carcinoma through the ZBTB16/TOP2A axis
Cell Death & Differentiation (2024)
-
Epigenetic and transcriptional regulation of cytokine production by Plasmodium falciparum-exposed monocytes
Scientific Reports (2024)
-
Transcriptomic screening of novel targets of sericin in human hepatocellular carcinoma cells
Scientific Reports (2024)
-
Echinococcus granulosus cyst fluid inhibits inflammatory responses through inducing histone demethylase KDM5B in macrophages
Parasites & Vectors (2023)
-
Epigenetics as a versatile regulator of fibrosis
Journal of Translational Medicine (2023)