Abstract
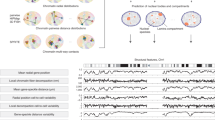
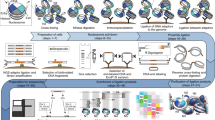
Chrom3D is a computational platform for 3D genome modeling that simulates the spatial positioning of chromosome domains relative to each other and relative to the nuclear periphery. In Chrom3D, chromosomes are modeled as chains of contiguous beads, in which each bead represents a genomic domain. In this protocol, a bead represents a topologically associated domain (TAD) mapped from ensemble Hi-C data. Chrom3D takes as input data significant pairwise TAD–TAD interactions determined from a Hi-C contact matrix, and TAD interactions with the nuclear periphery, determined by ChIP-sequencing of nuclear lamins to define lamina-associated domains (LADs). Chrom3D is based on Monte Carlo simulations initiated from a starting random bead configuration. During the optimization process, TAD–TAD interactions constrain bead positions relative to each other, whereas LAD information constrains the corresponding bead toward the nuclear periphery. Optimization can be repeated many times to generate an ensemble of 3D genome models. Analyses of the models enable estimations of the radial positioning of genomic sites in the nucleus across cells in a population. Chrom3D provides opportunities to reveal spatial relationships between TADs and LADs. More generally, predictions from Chrom3D models can be experimentally tested in the laboratory. We describe the entire Chrom3D protocol for modeling a 3D diploid human genome, from the creation of input files to the final rendering of 3D genome structures. The procedure takes ∼18 h. Chrom3D is freely available on GitHub.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Dekker, J., Marti-Renom, M.A. & Mirny, L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Guelen, L. et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008).
van Steensel, B. & Dekker, J. Genomics tools for unraveling chromosome architecture. Nat. Biotechnol. 28, 1089–1095 (2010).
Collas, P., Lund, E.G. & Oldenburg, A.R. Closing the (nuclear) envelope on the genome: how nuclear lamins interact with promoters and modulate gene expression. BioEssays 36, 75–83 (2014).
Lund, E.G., Duband-Goulet, I., Oldenburg, A., Buendia, B. & Collas, P. Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin. Nucleus 6, 30–38 (2015).
Gesson, K. et al. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 26, 462–473 (2016).
Robson, M.I. et al. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol. Cell 62, 834–847 (2016).
Robson, M.I. et al. Constrained release of lamina-associated enhancers and genes from the nuclear envelope during T-cell activation facilitates their association in chromosome compartments. Genome Res. 27, 1126–1138 (2017).
Dixon, J.R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E.P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Nagano, T. et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017).
Hansen, A.S., Pustova, I., Cattoglio, C., Tjian, R. & Darzacq, X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 6 e25776 (2017).
Barbieri, M. et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. USA 109, 16173–16178 (2012).
Bohn, M. & Heermann, D.W. Repulsive forces between looping chromosomes induce entropy-driven segregation. PLoS One 6, e14428 (2011).
Mateos-Langerak, J. et al. Spatially confined folding of chromatin in the interphase nucleus. Proc. Natl. Acad. Sci. USA 106, 3812–3817 (2009).
Rosa, A. & Everaers, R. Ring polymers in the melt state: the physics of crumpling. Phys. Rev. Lett. 112, 118302 (2014).
Vettorel, T., Grosberg, A.Y. & Kremer, K. Statistics of polymer rings in the melt: a numerical simulation study. Phys. Biol. 6, 025013 (2009).
Naumova, N. et al. Organization of the mitotic chromosome. Science 14, 953 (2013).
Cook, P.R. & Marenduzzo, D. Entropic organization of interphase chromosomes. J. Cell Biol. 186, 825–834 (2009).
Jerabek, H. & Heermann, D.W. Expression-dependent folding of interphase chromatin. PLoS One 7, e37525 (2012).
Kalhor, R., Tjong, H., Jayathilaka, N., Alber, F. & Chen, L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 30, 90–98 (2011).
Tjong, H. et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc. Natl. Acad. Sci. USA 113, E1663–E1672 (2016).
Paulsen, J. et al. Chrom3D: three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biol. 18, 21 (2017).
Perovanovic, J. et al. Laminopathies disrupt epigenomic developmental programs and cell fate. Sci. Transl. Med. 8, 335ra58 (2016).
Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet 2, 292–301 (2001).
Rao, S.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Kind, J. et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163, 134–147 (2015).
Lund, E. et al. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 23, 1580–1589 (2013).
Oldenburg, A. et al. A lipodystrophy-causing lamin A mutant alters conformation and epigenetic regulation of the anti-adipogenic MIR335 locus. J. Cell Biol. 216, 2731–2743 (2017).
Kind, J. et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153, 178–192 (2013).
Stevens, T.J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Garcia-Nieto, P.E. et al. Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 36, 2829–2843 (2017).
Bau, D. et al. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat. Struct. Mol. Biol. 18, 107–114 (2011).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012).
Serra, F. et al. Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS Lett. 589, 2987–2995 (2015).
Szalaj, P. et al. An integrated 3-dimensional genome modeling engine for data-driven simulation of spatial genome organization. Genome Res. 26, 1697–1709 (2016).
Russel, D. et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244 (2012).
Bau, D. & Marti-Renom, M.A. Genome structure determination via 3C-based data integration by the Integrative Modeling Platform. Methods 58, 300–306 (2012).
Serra, F. et al. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput. Biol. 13, e1005665 (2017).
Duan, Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367 (2010).
Li, Q. et al. The three-dimensional genome organization of Drosophila melanogaster through data integration. Genome Biol. 18, 145 (2017).
Dai, C. et al. Mining 3D genome structure populations identifies major factors governing the stability of regulatory communities. Nat. Commun. 7, 11549 (2016).
Lund, E.G., Oldenburg, A.R. & Collas, P. Enriched Domain Detector: a program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res. 42, e92 (2014).
Durand, N.C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Gundersen, S. et al. Identifying elemental genomic track types and representing them uniformly. BMC Bioinformatics 12, 494 (2011).
Yang, Z. et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 179, 269–278 (2012).
de Nooijer, S., Wellink, J., Mulder, B. & Bisseling, T. Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei. Nucleic Acids Res. 37, 3558–3568 (2009).
Lesne, A., Riposo, J., Roger, P., Cournac, A. & Mozziconacci, J. 3D genome reconstruction from chromosomal contacts. Nat. Methods 11, 1141–1143 (2014).
Rousseau, M., Fraser, J., Ferraiuolo, M.A., Dostie, J. & Blanchette, M. Three-dimensional modeling of chromatin structure from interaction frequency data using Markov chain Monte Carlo sampling. BMC Bioinformatics 12, 414–412 (2011).
Bau, D. & Marti-Renom, M.A. Structure determination of genomic domains by satisfaction of spatial restraints. Chromosome Res. 19, 25–35 (2011).
Mifsud, B. et al. GOTHiC, a probabilistic model to resolve complex biases and to identify real interactions in Hi-C data. PLoS One 12, e0174744 (2017).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
P.C. is supported by the Research Council of Norway, the Norwegian Cancer Society, South-East Health Norway, The Norwegian Center for Stem Cell Research and the University of Oslo.
Author information
Authors and Affiliations
Contributions
J.P. and T.M.L.A. wrote code, performed data analysis and wrote the manuscript. J.P. conceptualized and developed Chrom3D. P.C. supervised the work and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Paulsen, J., Liyakat Ali, T. & Collas, P. Computational 3D genome modeling using Chrom3D. Nat Protoc 13, 1137–1152 (2018). https://doi.org/10.1038/nprot.2018.009
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.009
This article is cited by
-
Near telomere-to-telomere genome of the model plant Physcomitrium patens
Nature Plants (2024)
-
DiffDomain enables identification of structurally reorganized topologically associating domains
Nature Communications (2024)
-
Strong interactions between highly dynamic lamina-associated domains and the nuclear envelope stabilize the 3D architecture of Drosophila interphase chromatin
Epigenetics & Chromatin (2023)
-
Chromatin domain alterations linked to 3D genome organization in a large cohort of schizophrenia and bipolar disorder brains
Nature Neuroscience (2022)
-
Time-dependent effect of 1,6-hexanediol on biomolecular condensates and 3D chromatin organization
Genome Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.