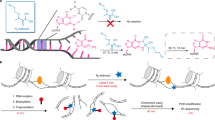
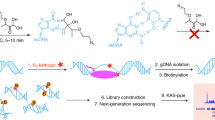
Abstract
Aberrant transcription induced by DNA damage may confer risk for the development of cancer and other human diseases. Traditional methods for measuring lesion-induced transcriptional alterations often involve extensive colony screening and DNA sequencing procedures. Here we describe a protocol for the quantitative assessment of the effects of DNA lesions on the efficiency and fidelity of transcription in vitro and in mammalian cells. The method is also amenable to investigating the influence of specific DNA repair proteins on the biological response toward DNA damage during transcription by manipulating their gene expression. Specifically, we present detailed, step-by-step procedures, including DNA template preparation, in vitro and in vivo transcription, RNA purification, reverse-transcription PCR (RT-PCR) and restriction digestion of RT-PCR products. Analyses of restriction fragments of interest are performed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) and polyacrylamide gel electrophoresis (PAGE). The entire procedure described in this protocol can be completed in 15–20 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bregeon, D. & Doetsch, P.W. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer 11, 218–227 (2011).
Morreall, J.F., Petrova, L. & Doetsch, P.W. Transcriptional mutagenesis and its potential roles in the etiology of cancer and bacterial antibiotic resistance. J. Cell Physiol. 228, 2257–2261 (2013).
Gaillard, H., Herrera-Moyano, E. & Aguilera, A. Transcription-associated genome instability. Chem. Rev. 113, 8638–8661 (2013).
Lange, S.S., Takata, K. & Wood, R.D. DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 (2011).
Sale, J.E., Lehmann, A.R. & Woodgate, R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 (2012).
Saxowsky, T.T. & Doetsch, P.W. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem. Rev. 106, 474–488 (2006).
Kuraoka, I. & Tanaka, K. Assays for transcription elongation by RNA polymerase II using oligo(dC)-tailed template with single DNA damage. Methods Enzymol. 408, 214–223 (2006).
You, C. et al. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 8, 817–822 (2012).
Delaney, J.C. & Essigmann, J.M. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 408, 1–15 (2006).
Frick, L.E., Delaney, J.C., Wong, C., Drennan, C.L. & Essigmann, J.M. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc. Natl. Acad. Sci. USA 104, 755–760 (2007).
Hong, H.Z., Cao, H.C. & Wang, Y.S. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 35, 7118–7127 (2007).
Yuan, B.F., Cao, H.C., Jiang, Y., Hong, H.Z. & Wang, Y.S. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. USA 105, 8679–8684 (2008).
You, C., Dai, X., Yuan, B. & Wang, Y. Effects of 6-thioguanine and S6-methylthioguanine on transcription in vitro and in human cells. J. Biol. Chem. 287, 40915–40923 (2012).
You, C., Wang, P., Dai, X. & Wang, Y. Transcriptional bypass of regioisomeric ethylated thymidine lesions by T7 RNA polymerase and human RNA polymerase II. Nucleic Acids Res. 42, 13706–13713 (2014).
You, C., Wang, J., Dai, X. & Wang, Y. Transcriptional inhibition and mutagenesis induced by N-nitroso compound-derived carboxymethylated thymidine adducts in DNA. Nucleic Acids Res. 43, 1012–1018 (2015).
You, C., Ji, D., Dai, X. & Wang, Y. Effects of Tet-mediated oxidation products of 5-methylcytosine on DNA transcription in vitro and in mammalian cells. Sci. Rep. 4, 7052 (2014).
Wang, J. & Wang, Y. Chemical synthesis of oligodeoxyribonucleotides containing N3- and O4-carboxymethylthymidine and their formation in DNA. Nucleic Acids Res. 37, 336–345 (2009).
Nagel, Z.D. et al. Multiplexed DNA repair assays for multiple lesions and multiple doses via transcription inhibition and transcriptional mutagenesis. Proc. Natl. Acad. Sci. USA 111, E1823–E1832 (2014).
Rizzo, J.M. & Buck, M.J. Key principles and clinical applications of 'next-generation' DNA sequencing. Cancer Prev. Res. 5, 887–900 (2012).
Baker, D. et al. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 282, 22592–22604 (2007).
Luhnsdorf, B. et al. Generation of reporter plasmids containing defined base modifications in the DNA strand of choice. Anal. Biochem. 425, 47–53 (2012).
Wang, H. & Hays, J.B. Simple and rapid preparation of gapped plasmid DNA for incorporation of oligomers containing specific DNA lesions. Mol. Biotechnol. 19, 133–140 (2001).
Kitsera, N. et al. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 39, 5926–5934 (2011).
Bregeon, D., Peignon, P.A. & Sarasin, A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 5, e1000577 (2009).
Bregeon, D. & Doetsch, P.W. Reliable method for generating double-stranded DNA vectors containing site-specific base modifications. BioTechniques 37, 760–762, 764, 766 (2004).
Enoiu, M., Ho, T.V., Long, D.T., Walter, J.C. & Scharer, O.D. Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol. Biol. 920, 203–219 (2012).
Edelheit, O., Hanukoglu, A. & Hanukoglu, I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 9, 61 (2009).
Bomgarden, R. et al. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 25, 5036–5036 (2006).
Chowdhury, G. & Guengerich, F. Liquid chromatography-mass spectrometry analysis of DNA polymerase reaction products. Curr. Protoc. Nucleic Acid Chem. 47, 7.16.1–7.16.11 (2011).
Acknowledgements
We acknowledge T.R. O'Connor (The City of Hope) and K.A. Cimprich (Stanford University) for kindly providing the initial pTGFP-Hha10 vector and human fibroblast cells (XP12RO and GM15876A), respectively. This work was supported by US National Institutes of Health (R01DK082779, R01ES019873, R01 ES025121 and R01CA101864 to Y.W.).
Author information
Authors and Affiliations
Contributions
C.Y. designed and performed the research and wrote the manuscript. Y.W. designed and supervised the research and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
You, C., Wang, Y. Quantitative measurement of transcriptional inhibition and mutagenesis induced by site-specifically incorporated DNA lesions in vitro and in vivo. Nat Protoc 10, 1389–1406 (2015). https://doi.org/10.1038/nprot.2015.094
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.094
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.