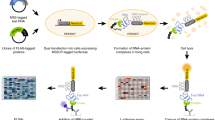
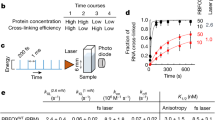
Abstract
RNAi is well known for its ability to regulate gene expression in the cytoplasm of mammalian cells. In mammalian cell nuclei, however, the impact of RNAi has remained more controversial. A key technical hurdle has been a lack of optimized protocols for the isolation and analysis of cell nuclei. Here we describe a simplified protocol for nuclei isolation from cultured cells that incorporates a method for obtaining nucleoplasmic and chromatin fractions and removing cytoplasmic contamination. Cell fractions can then be used to detect the presence and activity of RNAi factors in the nucleus. We include a method for investigating an early step in RNAi, Argonaute protein loading with small RNAs, which is enabled by our improved extract preparations. This protocol facilitates the characterization of nuclear RNAi, and it can be applied to the analysis of other nuclear proteins and pathways. From cellular fractionation to analysis of Argonaute loading results, this protocol takes 4–6 d to complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wilson, R.C. & Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42, 217–239 (2013).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Morris, K.V., Chan, S.W., Jacobsen, S.E. & Looney, D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305, 1289–1292 (2004).
Janowski, B.A. et al. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigen RNAs. Nat. Chem. Bio. 1, 216–222 (2005).
Castanotto, D. et al. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol. Ther. 12, 179–183 (2005).
Ting, A.H., Schuebel, K.E., Herman, J.G. & Baylin, S.B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 37, 906–910 (2005).
Napoli, S., Pastori, C., Magistri, M., Carbone, G.M. & Catapano, C.V. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 28, 1708–1719 (2009).
Kim, D.H., Villeneuve, L.M., Morris, K.V. & Rossi, J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 13, 793–797 (2006).
Li, L.C. et al. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA 103, 17337–17342 (2006).
Janowski, B.A. et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 3, 166–173 (2007).
Matsui, M. et al. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 41, 10086–10109 (2013).
Huang, V. et al. RNAa is conserved in mammalian cells. PLoS ONE 5, e8848 (2010).
Liu, J., Hu, J. & Corey, D.R. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic Acids Res. 40, 1240–1250 (2012).
Allo, M. et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16, 717–724 (2009).
Janowski, B.A. et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 13, 787–792 (2006).
Chu, Y., Yue, X., Younger, S.T., Janowski, B.A. & Corey, D.R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 38, 7736–7748 (2010).
Vickers, T.A. et al. Efficient reduction of target RNAs by small interfering RNA and RNase H–dependent antisense agents: a comparative analysis. J. Biol. Chem. 278, 7108–7118 (2003).
Zeng, Y. & Cullen, B.R. RNA interference in human cells is restricted to the cytoplasm. RNA 8, 855–860 (2002).
Ikeda, K. et al. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using monoclonal antibody. J. Immunol. Methods 317, 38–44 (2006).
Stalder, L. et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 32, 1115–1127 (2013).
Ando, Y. et al. Nuclear pore complex protein mediated nuclear localization of dicer protein in human cells. PLoS ONE 6, E23385 (2011).
Doyle, M. et al. The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA 19, 1238–1252 (2013).
Ohrt, T., Muetze, J., Svoboda, P. & Schwille, P. Intracellular localization and routing of miRNA and RNAi pathway components. Curr. Top. Med. Chem. 12, 79–88 (2012).
Rudel, S., Flatley, A., Weinmann, L., Kremmer, E. & Meister, G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA 14, 1244–1253 (2008).
Till, S. et al. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 14, 897–903 (2007).
Weinmann, L. et al. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136, 496–507 (2009).
Gagnon, K.T., Li, L., Chu, Y., Janowski, B.A. & Corey, D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 6, 211–221 (2014).
Holding, C. RNAi active in the nucleus? The Scientist http://www.the-scientist.com/?articles.view/articleNo/23231/title/RNAi-active-in-the-nucleus-/ (2005).
Hetzer, M.W. The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2, a000539 (2010).
Michelson, U. & von Hagen, J. Isolation of subcellular organelles and structures. Methods Enzymol. 463, 306–328 (2009).
Greenberg, M.E. & Bender, T.P. Identification of newly transcribed RNA. Curr. Protoc. Mol. Biol. 78, 4.10.11–14.10.12 (2007).
Acknowledgements
Funding was provided by the US National Institutes of Health (1F32HD060377/KTG, GM 73042/DRC and GM85080/BAJ), the Welch Foundation (I-1244/DRC) and the Cancer Prevention and Research Institute of Texas (RP120311/BAJ).
Author information
Authors and Affiliations
Contributions
K.T.G. and L.L. designed and performed the experiments, including optimization of subcellular fractionation and development of the in vitro Argonaute loading assay. K.T.G., L.L., B.A.J. and D.R.C. all participated in the writing of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Gagnon, K., Li, L., Janowski, B. et al. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc 9, 2045–2060 (2014). https://doi.org/10.1038/nprot.2014.135
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.135
This article is cited by
-
Cytotoxic activities of alkaloid constituents from the climbing stems and rhizomes of Sinomenium acutum against cancer stem cells
Journal of Natural Medicines (2024)
-
ASPSCR1-TFE3 reprograms transcription by organizing enhancer loops around hexameric VCP/p97
Nature Communications (2024)
-
PSMC3 promotes RNAi by maintaining AGO2 stability through USP14
Cellular & Molecular Biology Letters (2022)
-
Non-canonical role of wild-type SEC23B in the cellular stress response pathway
Cell Death & Disease (2021)
-
Identification and characterization of a novel Epstein-Barr Virus-encoded circular RNA from LMP-2 Gene
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.