Abstract
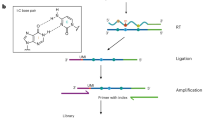
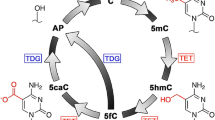
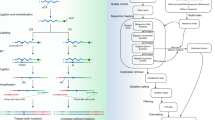
We describe a method for the efficient and selective identification of DNA containing the 5-hydroxymethylcytosine (5-hmC) modification. This protocol takes advantage of two proteins: T4 β-glucosyltransferase (β-gt), which converts 5-hmC to β-glucosyl-5-hmC (β-glu-5-hmC), and J-binding protein 1 (JBP1), which specifically recognizes and binds to β-glu-5-hmC. We describe the steps necessary to purify JBP1 and modify this protein such that it can be fixed to magnetic beads. Thereafter, we detail how to use the JBP1 magnetic beads to obtain DNA that is enriched with 5-hmC. This method is likely to produce results similar to those of other 5-hmC pull-down assays; however, all necessary components for the completion of this protocol are readily available or can be easily and rapidly synthesized using basic molecular biology techniques. This protocol can be completed in less than 2 weeks and allows the user to isolate 5-hmC-containing genomic DNA that is suitable for analysis by quantitative PCR (qPCR), sequencing, microarray and other molecular biology assays.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Penn, N.W. Modification of brain deoxyribonucleic acid base content with maturation in normal and malnourished rats. Biochem. J. 155, 709–712 (1976).
Cannon-Carlson, S.V., Gokhale, H. & Teebor, G.W. Purification and characterization of 5-hydroxymethyluracil-DNA glycosylase from calf thymus. Its possible role in the maintenance of methylated cytosine residues. J. Biol. Chem. 264, 13306–13312 (1989).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Ito, S. et al. Role of Tet proteins in 5mC to 5-hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
Szwagierczak, A., Bultmann, S., Schmidt, C.S., Spada, F. & Leonhardt, H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 38, e181 (2010).
Ko, M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010).
Guo, J.U., Su, Y., Zhong, C., Ming, G.L. & Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011).
Wu, H. et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684 (2011).
Wu, H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 (2011).
Robertson, J., Robertson, A.B. & Klungland, A. The presence of 5-hydroxymethylcytosine at the gene promoter and not in the gene body negatively regulates gene expression. Biochem. Biophys. Res. Commun. 411, 40–43 (2011).
Georgopoulos, C.P. & Revel, H.R. Studies with glucosyl transferase mutants of the T-even bacteriophages. Virology 44, 271–285 (1971).
Kornberg, S.R., Zimmerman, S.B. & Kornberg, A. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. J. Biol. Chem. 236, 1487–1493 (1961).
Gommers-Ampt, J.H. et al. β-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell 75, 1129–1136 (1993).
Borst, P. & Sabatini, R. Base J: discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 62, 235–251 (2008).
van Leeuwen, F. et al. β-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. USA 95, 2366–2371 (1998).
Sabatini, R., Meeuwenoord, N., van Boom, J.H. & Borst, P. Recognition of base J in duplex DNA by J-binding protein. J. Biol. Chem. 277, 958–966 (2002).
Cross, M. et al. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J. 18, 6573–6581 (1999).
Grover, R.K. et al. O-glycoside orientation is an essential aspect of base J recognition by the kinetoplastid DNA-binding protein JBP1. Angew Chem. Int. Ed. Eng. 46, 2839–2843 (2007).
Robertson, A.B. et al. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 39, e55 (2011).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011).
Stroud, H., Feng, S., Morey Kinney, S., Pradhan, S. & Jacobsen, S.E. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 12, R54 (2011).
Pastor, W.A. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 (2011).
Flusberg, B.A. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010).
Song, C.X., Yu, M., Dai, Q. & He, C. Detection of 5-hydroxymethylcytosine in a combined glycosylation restriction analysis (CGRA) using restriction enzyme Taq(α)I. Bioorg. Med. Chem. Lett. 21, 5075–5077 (2011).
Xu, S.Y., Corvaglia, A.R., Chan, S.H., Zheng, Y. & Linder, P. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res. 39, 5597–5610 (2011).
Szwagierczak, A. et al. Characterization of PvuRts1I endonuclease as a tool to investigate genomic 5-hydroxymethylcytosine. Nucleic Acids Res. 39, 5149–5156 (2011).
Song, C.X. et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 29, 68–72 (2011).
Nestor, C., Ruzov, A., Meehan, R. & Dunican, D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques 48, 317–319 (2010).
Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Acknowledgements
This work was funded by the Norwegian Research Council and the Norwegian Cancer Society.
Author information
Authors and Affiliations
Contributions
A.B.R., J.A.D. and A.K. initiated the project. A.B.R., J.A.D. and A.K. designed the study. A.B.R., J.A.D. and R.O. designed and conducted the experiments. A.B.R. drafted the manuscript and all the authors participated in writing and editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have been given a provisional patent for the method described in this protocol.
Supplementary information
Supplementary Note 1
Test oligonucleotide sequences and qPCR primers (DOC 31 kb)
Supplementary Note 2
Sequence of pET28a-JBP1 (DOC 42 kb)
Rights and permissions
About this article
Cite this article
Robertson, A., Dahl, J., Ougland, R. et al. Pull-down of 5-hydroxymethylcytosine DNA using JBP1-coated magnetic beads. Nat Protoc 7, 340–350 (2012). https://doi.org/10.1038/nprot.2011.443
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.443
This article is cited by
-
Metal-ion-induced DNAzyme on magnetic beads for detection of lead(II) by using rolling circle amplification, glucose oxidase, and readout of pH changes
Microchimica Acta (2019)
-
Mapping and elucidating the function of modified bases in DNA
Nature Reviews Chemistry (2017)
-
RRHP: a tag-based approach for 5-hydroxymethylcytosine mapping at single-site resolution
Genome Biology (2014)
-
Advances in the profiling of DNA modifications: cytosine methylation and beyond
Nature Reviews Genetics (2014)
-
DNA methylation in schizophrenia: progress and challenges of epigenetic studies
Genome Medicine (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.