Abstract
Protein expression abnormalities have been implicated in the pathophysiology of schizophrenia, but the underlying cause of these changes is not known. We sought to investigate ubiquitin and ubiquitin-like (UBL) systems (SUMOylation, NEDD8ylation, and Ufmylation) as putative mechanisms underlying protein expression abnormalities seen in schizophrenia. For this, we performed western blot analysis of total ubiquitination, free ubiquitin, K48- and K63-linked ubiquitination, and E1 activases, E2 conjugases, and E3 ligases involved in ubiquitination and UBL post-translational modifications in postmortem brain tissue samples from persons with schizophrenia (n=13) and comparison subjects (n=13). We studied the superior temporal gyrus (STG) of subjects from the Mount Sinai Medical Center brain collection that were matched for age, tissue pH, and sex. We found an overall reduction of protein ubiquitination, free ubiquitin, K48-linked ubiquitination, and increased K63 polyubiquitination in schizophrenia. Ubiquitin E1 activase UBA (ubiquitin activating enzyme)-6 and E3 ligase Nedd (neural precursor cell-expressed developmentally downregulated)-4 were decreased in this illness, as were E3 ligases involved in Ufmylation (UFL1) and SUMOylation (protein inhibitor of activated STAT 3, PIAS3). NEDD8ylation was also dysregulated in schizophrenia, with decreased levels of the E1 activase UBA3 and the E3 ligase Rnf7. This study of ubiquitin and UBL systems in schizophrenia found abnormalities of ubiquitination, Ufmylation, SUMOylation, and NEDD8ylation in the STG in this disorder. These results suggest a novel approach to the understanding of schizophrenia pathophysiology, where a disruption in homeostatic adaptation of the cell underlies discreet changes seen at the protein level in this illness.
Similar content being viewed by others
INTRODUCTION
Schizophrenia, a debilitating illness affecting 1% of the population, has a complex pathophysiology that is still not completely understood. Until recently, efforts have been made to unveil the neuropathology of this illness at the molecular level. Numerous studies have found decreased expression of multiple proteins in schizophrenia, including but not limited to dysbindin (Talbot et al, 2004; Weickert et al, 2008), cdc42 (Rubio et al, 2012), kalirin (Rubio et al, 2012), Akt1 (Zheng et al, 2012), Reelin and GAD67 (Guidotti et al, 2000), Rap2 (Funk et al, 2012), JNK1 (Funk et al, 2012), JNK2, (Funk et al, 2012), and PSD-95 (Funk et al, 2012). In addition, vesicle-sorting defects (Larimore et al, 2011) and decreased expression of proteins involved in forward trafficking of glutamate receptors have also been reported (Hammond et al, 2011).
The ubiquitin proteasome system (UPS), a protein degradation system, has been identified on the basis of genetic reports as a canonical pathway associated with neuropsychiatric disorders, including Alzheimer’s (Lam et al, 2000), Parkinson’s (Shimura et al, 2001), psychosis, and bipolar disorder (Bousman et al, 2010a, 2010b; Middleton et al, 2002). In addition to targeting proteins for degradation, the UPS is involved in vesicle trafficking, trafficking of proteins from the endoplasmic reticulum to the plasma membrane, recycling of receptors at the cell membrane (Haas and Broadie, 2008), and sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway (Scheuring et al, 2012). In addition, clathrin-coated vesicle trafficking interacting proteins have been found disrupted in schizophrenia. This is relevant as the clathrin interactome regulates trafficking and targeting of ubiquitinated proteins (Schubert et al, 2012). Owing to the overlapping nature of the UPS function with abnormalities found in schizophrenia, the proteasome system emerged as a putative candidate underlying protein changes in this illness. As such, abnormalities of the UPS have been repeatedly reported in mRNA expression studies performed in blood cells (Bousman et al, 2010a), hippocampus (Altar et al, 2005), prefrontal cortex, and temporal cortex (Aston et al, 2004; Vawter et al, 2001) of patients suffering from schizophrenia. In this study, we sought to determine whether post-translational mechanisms regulating protein degradation could be driving the abnormal protein expression pattern seen in schizophrenia. Because UPS and ubiquitin-like (UBL) systems regulate protein expression, trafficking, and sorting, we hypothesized that abnormalities in these pathways are likely to underlie the numerous protein expression disruptions that have been described in schizophrenia.
Protein ubiquitination is a three step process (Figure 1). Initially, ubiquitin is activated by E1 activases in an ATP-dependent process, resulting in a thiol ester intermediate, E1-S-ubiquitin. This high-energy compound is then transferred to a target substrate by an E2 conjugase, also forming a high-energy intermediate, E2-S-ubiquitin. The substrate is bound to specific E3 ligases that catalyze the final process by covalently binding ubiquitin to an internal lysine residue. During this step, the third and last high-energy thiol ester intermediate, ubiquitin-S-E3, is formed (Varshavsky, 2012). By adding activated ubiquitin residues to the internal lysine, a polyubiquitin chain is synthesized. The lysine residue at which polyubiquitination takes place determines the fate of the target protein. When polyubiquitination occurs at lysine 48 (K48), the protein is likely to be recognized by the 26S proteasome and degraded; polyubiquitin linkage at lysine 63 (K63) is known to have non-proteolytic functions associated with protein–protein interactions, cellular signaling, and endocytosis (Varshavsky, 2012). To date, there are at least 35 E2s and over 600 E3s that have been described in mammals, resulting in a system with a high degree of substrate specificity.
Ubiquitin and ubiquitin-like (UBL) pathway.
In addition to ubiquitin, several other proteins termed UBL modifiers have recently been described. The UBLs share little sequence homology with ubiquitin but have similar 3D structures, with a ubiquitin fold and a C-terminal glycine that allows them to bind to lysine residues of substrates in a UBL fashion. Among them, NEDD8 (neural precursor cell-expressed developmentally downregulated-8) regulates ubiquitin E3 ligases and leads to proteasome degradation. Ufm1 (ubiquitin-fold modifier-1) is involved in the prevention of ER stress-induced apoptosis (Lemaire et al, 2011); and SUMO (small ubiquitin-like modifier) is implicated in modulation of the activity of voltage-gated K+ channels (Plant et al, 2011), repair of DNA single-strand breaks in neurons (Hudson et al, 2012; Mao et al, 2000), axonal mRNA trafficking (Kadare et al, 2003), and neurodevelopment. NEDD8ylation dysfunction has been described in several degenerative disorders including Parkinson’s disease (Choo et al, 2012) and Alzheimer’s disease (Kee et al, 2012). Similarly, abnormalities of SUMOylation have been implicated in neurodegenerative disorders including Parkinson’s disease, Alzheimer’s disease, amyotrophic lateral sclerosis, spinocerebellar ataxia, and Huntington’s disease (Krumova and Weishaupt, 2012).
To assess the involvement of the ubiquitin and UBL systems in schizophrenia, we performed a detailed study including measurement of total lysine 48 and 63 protein ubiquitination, polyubiquitination, and a series of molecules involved in activation, conjugation, and ligation of ubiquitin in the left superior temporal gyrus (STG) in schizophrenia. The left STG participates in the development of auditory hallucinations (Barta et al, 1990; Nenadic et al, 2010; Silbersweig et al, 1995) and the volume of this cortical area is decreased in subjects with schizophrenia (Sun et al, 2009). In addition, the STG has been associated with thought process abnormalites seen in this illness (Rajarethinam et al, 2000). Several reports have found abnormal expression of ubiquitination-related transcripts (Aston et al, 2004; Vawter et al, 2001), and the density of GABAA receptors is increased (Deng and Huang, 2006) in the STG in schizophrenia. We found decreased protein ubiquitination, accompanied by decreased ubiquitin, and UBL activases and ligases, suggesting the possibility that disruption of these systems may underlie protein expression abnormalities found in postmortem studies in schizophrenia.
METHODS AND MATERIALS
Subjects, Tissue Acquisition and Preparation
Samples from the STG were obtained for thirteen closely matched subjects (schizophrenia and comparison) from the Mount Sinai Medical Center brain collection (Table 1). Patients diagnosed with schizophrenia by two independent clinicians using DSM-III-R criteria were recruited prospectively (Bauer et al, 2008). Each patient had a documented history of psychotic symptoms before the age of 40 and at least 10 years of hospitalization. The subjects were evaluated for vascular dementia, probable or possible diffuse Lewy body disease, Parkinson’s disease, frontotemporal dementia, drug, or alcohol abuse, and underwent cognition tests including the MMSE and CDR. In addition, each brain was examined neuropathologically by systematized macroscopic and microscopic evaluation using CERAD guidelines. The subjects studied did not show neuropathological evidence to meet the criteria for neurodegenerative disorders including Alzheimer’s disease (Purohit et al, 1998). Comparison subjects were evaluated for and free from psychiatric illnesses, history of substance abuse, and neurodegenerative disorders. Exclusion criteria included substance abuse, suicide, or coma for more than 6 h before death. Tobacco use data were not consistently available for all subjects. Next of kin consent to perform an autopsy on the body and brain for diagnostic and research purposes was obtained for each subject.
Brain dissection was limited to 0.8–1 cm3 of gray matter of the left STG cortical region. The dissected tissue was immediately frozen at −80 °C until use. Samples were then homogenized using a Power Gen 125 homogenizer (Thermo Fisher Scientific, Rockford, Illinois) in RIPA buffer consisting of 50 mM Tris HCl (pH 7.4), 150 mM NaCl, 0.5 mM EGTA, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.5% SDS, deubiquitinase inhibitors (5 mM N-ethylmaleimide, 50 mM iodoacetamide), and a protease and a phosphatase inhibitor tablet (Complete Mini and Phostop, respectively, Roche Diagnostics, Manheim, Germany). Homogenates were assayed for protein concentration using a BCA protein assay kit (Thermo Fisher Scientific, Rockford, Illinois), aliquoted, and stored at −80 °C until use.
Subjects were matched for age, sex, and tissue pH (Table 1). In addition, we attempted to also match for PMI although these were not as closely matched.
Rodent Antipsychotic Drug Treatment Experiments
Male Sprague-Dawley rats (250 g) were housed in pairs and injected intramuscularly every 3 weeks, for a total of 12 injections, with vehicle (sesame oil) or 28.5 mg/kg of haloperidol decanoate in sesame oil. Rats were decapitated, the brains were immediately collected, the left anterior cortex was dissected on wet ice, and was prepared as described above. Ten haloperidol-treated and ten vehicle-injected rats were used for each experiment. All experiments were carried out according to UAB IACUC guidelines and regulations.
Western Blot Analysis
Western blot analyses were performed as previously described (Rubio et al, 2012). Samples were diluted in ultrapure water and a reducing buffer to a concentration of 20 μg protein per 10 μl and denatured at 70 °C for 10 min. For each subject, 20 μg total protein per lane was loaded in duplicate into a 4–12% gradient polyacrylamide bis-tris gel (Invitrogen, Carlsbad, California). For each gel, comparison and schizophrenia samples were loaded into wells in alternating lanes. The antibody concentration was optimized for each protein to ensure that detection was within the linear range of the assay and that the primary antibody was present in excess.
After electrophoresis, samples were transferred to PVDF membranes using Bio-Rad semi-dry transblotters (Hercules, California). Membranes were blocked with Li-Cor blocking buffer (Lincoln, Nebraska) or 5% bovine serum albumin (BSA) in phosphate-buffered solution (PBS) for 1 h at room temperature. Blots were then probed with primary antisera diluted either in Li-Cor blocking buffer containing 0.1% Tween-20 or 5% BSA buffer containing 0.1% Tween-20 (Supplementary Table S1). Blots were incubated overnight at 4 °C except for VCP and β-tubulin, which were incubated for 1 h at room temperature. Membranes were washed twice for 10 min in PBS and probed for 1 h at room temperature with goat anti-mouse, goat anti-rabbit or rabbit anti-goat IR-Dye 670 or 800cw-labeled secondary antibody diluted in Li-Cor and 0.1% Tween-20, or 5% BSA buffer and 0.1% Tween-20. After two 10-min washes in PBS, membranes were imaged using a Li-Cor Odyssey scanner. Loading control proteins (VCP, 110 kDa and β-tubulin, 55 kDa) were chosen for each assay based on the molecular weight of the target protein to avoid potential interference based on similar sizes. There were no significant differences in either VCP or β-tubulin expression between diagnostic groups, consistent with a previous report from the STG (Moehle et al, 2012). Full-blot images for all proteins studied are shown in Supplementary Figure S1.
Data Analysis
For each assay, boxes were manually placed around each band of expected molecular weight to obtain integrated intensity values using Odyssey 3.0 analytical software (Li-Cor, Lincoln, Nebraska). For total ubiquitin studies, a box was manually outlined around each lane to obtain the integrated intensity value of each individual lane. Intra-lane background was subtracted. The value of each band or lane of interest was normalized to the in-lane value of β-tubulin or VCP. For each subject, duplicate normalized data were averaged and the resulting values were used for statistical analysis. Data were analyzed using Statistica software (Statsoft, Tulsa, Oklahoma). All data were normally distributed. Correlation analyses were performed to determine associations between the dependent variables and tissue pH, age, and PMI. No correlation with these factors were seen, and one-way analysis of variance was used as the primary statistical test. Although PMI was not correlated with any dependent measure, we also performed ANCOVA with PMI as a covariate as a secondary post-hoc test; no results were changed when data were analyzed in this manner. For rat experiments, unpaired two-tailed t-tests were used. For all tests α=0.05.
RESULTS
Free Ubiquitin, Total Ubiquitination, K48 and K63-Linked Polyubiquitination are Abnormal in Schizophrenia
To test if protein ubiquitination is affected in schizophrenia, we first measured total protein ubiquitination in the STG of schizophrenia and control subjects. We found that total ubiquitination (Figure 2a) (F(1,23)=5.2, p=0.03) and free ubiquitin (Figure 2b) (F(1,24)=4.5, p=0.04) are decreased in schizophrenia. We then explored the possibility that polyubiquitin linkage at specific lysine residues may be affected in schizophrenia. We analyzed the integrated intensity of each individual band for K48 and K63-linked polyubiquitination. A detailed analysis showed decreased K48-linked polyubiquitination in 70 and 42 kDa proteins (bands 3 and 6, Figure 2c) (F(1,22)=6.5, p=0.02; F(1,24)=6.8, p=0.02), and increased K63-linked polyubiquitination of 40 kDa proteins (band 5, Figure 2d) (F(1,24)=5.8, p=0.02) were found in schizophrenia.
Overall protein ubiquitination is decreased in schizophrenia. Protein expression of total ubiquitin (a) and free ubiquitin (b) were found decreased in schizophrenia. Analysis of individual western blot bands shows a decrease in K48-linked ubiquitination (c) band shown corresponds to a comparison subject) and an increase of K63-linked ubiquitination (d) band shown corresponds to a comparison subject) in the superior temporal gyrus (STG). Data are expressed as a ratio of the optical density value for the protein of interest/the optical density of VCP from the same patient. C, comparison; S, schizophrenia; VCP, valosin-containing protein. *p<0.05.
E1 Activases Involved in Ubiquitination are Decreased in Schizophrenia
Because abnormalities in intermediate enzymes involved in ubiquitination may lead to dysregulation of this post-translational pathway, we sought to determine if E1, E2, and E3 proteins are abnormally expressed in the STG in schizophrenia. Previous studies in peripheral blood (Bousman et al, 2010a, 2010b) and postmortem hippocampus (Altar et al, 2005) found decreased mRNA encoding E1 ubiquitin activases, therefore, we analyzed protein expression levels of the E1 activases UBA (ubiquitin activating enzyme)-1 and UBA6 (ubiquitin activating enzyme 6). Although UBA1 expression did not differ between schizophrenia and comparison subjects (Figure 3a) (F(1,24)=3.6), UBA6 expression (Figure 3b) (F(1,22)=5.3, p=0.03) was decreased in schizophrenia.
E1 activase UBA (ubiquitin activating enzyme)-6 and E3 ligase Nedd4 (neural precursor cell-expressed developmentally downregulated 4) involved in protein ubiquitination are decreased in schizophrenia in the superior temporal gyrus (STG). E1 UBA1 (a) is not altered in schizophrenia, whereas UBA6 (b) shows a significant decrease in this illness. E2 UBE2K (c) remains unchanged. Of the E3s studied, only Nedd4 (d) is abnormal in schizophrenia. SIAH2 (seven in absentia homolog 2) (e) ZINF1 (f), MIB2 (g) and TRIM62 (h) protein expression levels are comparable to the comparison group. Data are expressed as a ratio of the optical density value for the protein of interest/the optical density of VCP or β-tubulin from the same patient. C, comparison; S, schizophrenia; VCP, valosin-containing protein. *p<0.05.
E2 Conjugase UBE2K Involved in Ubiquitination is Not Changed in Schizophrenia
E2 conjugase UBE2K mRNA levels in lymphocytes are correlated with positive symptoms of schizophrenia (Bousman et al, 2010a), therefore, we sought to explore the possibility that this protein is abnormally expressed in STG. Our analysis showed no change in UBE2K protein levels in this illness (Figure 3c) (F(1,26)=0.6).
Expression of the E3 Ligase Nedd4 is Decreased in Schizophrenia
We measured the expression of five E3 ligases involved in ubiquitination in the STG of schizophrenia and comparison subjects. Of the five E3 ligases studied, Nedd4 (neural precursor cell-expressed, developmentally downregulated 4, (Figure 3d) (F(1,24)=4.3, p=0.04) was decreased in this illness. The E3 ligases SIAH2 (seven in absentia homolog 2, Figure 3e) (F(1,26)=0.8), whose mRNA levels in lymphocytes are correlated with positive symptoms of schizophrenia (Bousman et al, 2010b); ZNRF1 (Figure 3f) (F(1,26)=0.4), involved in establishment and maintenance of synaptic transmission and plasticity; MIB2 (Figure 3g) (F(1,18)=2), which regulates a candidate susceptibility gene for schizophrenia, NOTCH; and TRIM62 (Figure 3h) (F(1,24)=2.5) were not changed in schizophrenia, likely reflecting abnormalities of specific ligases in this illness.
UBL Protein Modifier Systems are Abnormal in Schizophrenia
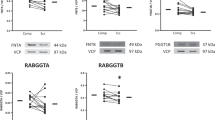
UBLs are recently characterized proteins involved in non-proteolytic processes such as anterograde and retrograde transport, and transcriptional regulation(van der Veen and Ploegh, 2012). They covalently attach to proteins similarly to ubiquitin. Among the UBLs, Ufm1 interacts with proteins through specific E1, E2, and E3-like substrates, namely Uba5, Ufc1, and Ufl1, resulting in Ufmylation of the substrate. We found that although Uba5 (Figure 4a) (F(1,24)=0.1) and Ufc1 (Figure 4b) (F(1,24)=1.7) were unchanged, Ufl1 was decreased in schizophrenia (Figure 4c) (F(1,23)=4.7, p=0.04) in the STG.
Ufl1, an E3 ligase involved in Ufmylation, is decreased in schizophrenia. E1 UBA (ubiquitin activating enzyme)-5 (a) and E2 Ufc1 (b) are not altered in schizophrenia, whereas UFL1 (c), the only known ligase involved in Ufmylation, shows a significant decrease in protein expression. Data are expressed as a ratio of the optical density value for the protein of interest/the optical density of VCP from the same patient. C, comparison; S, schizophrenia; VCP, valosin-containing protein. *p<0.05.
The UBL protein NEDD8 covalently binds to cullin subunits of E3 ligases, enhancing the attachment of E2 conjugases and thus protein ubiquitination (Kawakami et al, 2001). We found that one of the E1 proteins comprising the heterodimer involved in NEDD8ylation, formed by UBA3 (Figure 5a) (F(1,22)=5.3, p=0.03) and NAE1 (NEDD8-activating enzyme 1, Figure 5b) (F(1,24)=3.1793) is decreased in schizophrenia in the STG as is the E3 ligase RNF7 (Figure 5c) (F(1,23)=5.4, p=0.03), but not Rbx1 (Figure 5d) (F(1, 24)=0.6).
Nedd8ylation is disrupted in schizophrenia. UBA (ubiquitin activating enzyme)-3 (a), an E1 activases involved in Nedd8ylation, is significantly decreased in schizophrenia, whereas NAE1 (b) is not. Of the E3 ligases studied, only RNF7 (c) was found decreased, whereas Rbx(d) remained unchanged when compared with a comparison group. Data are expressed as a ratio of the optical density value for the protein of interest/the optical density of VCP from the same patient. C, comparison; S, schizophrenia; VCP, valosin-containing protein. *p<0.05.
We also studied proteins involved in SUMOylation, an UBL pathway involved in diverse cellular functions such as activation, translocation, and trafficking of proteins(Geiss-Friedlander and Melchior, 2007). SAE1 (SUMO-activating enzyme 1) and UBA2 (also known as SAE2) form a E1 heterodimer that activates the SUMO (small ubiquitin-related modifier) enzyme, which is then accepted by the E2 conjugase Ubc9 and linked to target proteins through specific E3 ligases (Melchior, 2000). We found that neither of the proteins associated with the E1 SAE1/UBA2 complex (Figures 6a and b) (F(1,24)=0.1; F(1,24)=3.3) nor the E2 SUMO pathway conjugase Ubc9 (Figure 6c) (F(1,24)=2) were affected in schizophrenia. Of the E3 ligases studied that are involved in this pathway in the STG, we found that PIAS3 (protein inhibitor of activated STAT, 3) was decreased in schizophrenia (Figure 6d) (F(1,23)=7.5, p=0.01), whereas TOPORS (topoisomerase I binding, arginine/serine rich, Figure 6e) (F(1,24)=0.6) and PIAS4 (protein inhibitor of activated STAT 4; Figure 6f) (F(1,24)=0.2) were unchanged.
PIAS3 (protein inhibitor of activated STAT3), an E3 ligase involved in SUMOylation, is decreased in schizophrenia. Analysis of optical densities of E1 activases SAE1 (SUMO-activating enzyme 1) (a) and UBA (ubiquitin activating enzyme)-2 (b), which form a heteromeric complex, showed no differences in protein expression in schizophrenia when compared with a comparison group. SUMO pathway E2 conjugase Ubc9 (c) also remains unchanged in schizophrenia. Of the E3 ligases studied, PIAS3 (d) showed a significant decrease, whereas TOPORS (topoisomerase I binding, arginine/serine rich) PIAS4 (protein inhibitor of activated STAT, 4) and PIAS1 (Protein inhibitor of activated STAT 1) remained unchanged (e, f, g). Data are expressed as a ratio of the optical density value for the protein of interest/the optical density of VCP or tubulin from the same patient. C, comparison; S, schizophrenia; VCP, valosin-containing protein. *p<0.05.
Finally, we measured ATG7 (autophagy-related protein 7) and MOCS3 (molybdenum cofactor synthesis 3), E1s involved in the activation of ATG8 (autophagy-related protein 8) and ATG12 (autophagy-related protein 12) families, and ubiquitin-related modifier 1 (URM1). ATG7 (Figure 7a) (F(1,22)=5.1, p=0.03) and MOCS3 (also known as UBA4, Figure 7b) (F (1,23)=4.8, p=0.04) were both decreased in schizophrenia in the STG. These data are summarized in Table 2.
E1 involved in ubiquitin-like (UBL) pathways are decreased in schizophrenia. ATG7 (autophagy-related protein 7) (a) and MOCS3 (molybdenum cofactor synthesis 3) (b) are E1s involved in UBL pathways that do not have a proteolytic role. Both of them were found significantly decreased in schizophrenia in the superior temporal gyrus (STG). Data are expressed as a ratio of the optical density value for the protein of interest/the optical density of VCP from the same patient. C, comparison; S, schizophrenia; VCP, valosin-containing protein. *p<0.05.
Chronic Treatment with Haloperidol Does Not Affect Ubiquitination or UBL Pathways
As most of the schizophrenia subjects studied were receiving chronic antipsychotic treatment either at or shortly before the time of death, we sought to address the possibility that the changes in the proteins involved in ubiquitin and UBL pathways that we observed could be due to medication effects. We measured protein expression of the UPS and UBL system in the brains of rats chronically treated with haloperidol. We found no differences in protein expression of any of the proteins that we found altered in the human experiments (Supplementary Figures S2 and S3). To further address the possibility of a treatment effect on protein expression levels, we performed post-hoc comparisons of protein expression levels of patients that were on or off antipsychotic medication for at least 6 weeks before death, and found no differences between these subgroups, although such an analysis is limited by the small sample size.
DISCUSSION
This study of ubiquitin and UBL systems in the STG found abnormal protein ubiquitination in schizophrenia, including decreased-free ubiquitin and abnormalities of E1 activases and E3 ligases. In addition, we also found abnormal expression of E1 and E3s involved in SUMOylation, Nedd8ylation, and Ufmylation. Previous reports support these findings, including decreased mRNA expression of genes involved in the UPS in the prefrontal cortex (Middleton et al, 2002; Vawter et al, 2001) and dentate gyrus (Altar et al, 2005). In a recent peripheral blood microarray study, the ubiquitin pathway was found to be one of the top 10 canonical pathways disrupted in schizophrenia (Bousman et al, 2010a, 2010b).
Characteristics of schizophrenia include abnormalities of synapses and synaptic proteins (Yin et al, 2012). The number of dendritic spines and several protein pathways involved in their development and maintenance are abnormal in this illness (Penzes et al, 2011). Dendritic spine structure is dependent on the coordinated balance between protein synthesis and degradation. Therefore, it is probable that the UPS system underlies some aspects of synaptic dysfunction in schizophrenia. Our findings have expanded previous reports by providing a possible mechanism by which ubiquitination is altered in this illness. We found a decrease in one of the E1s involved in ubiquitination, UBA6, which interacts specifically with the E2 USE1. A recent study in mice found that CRE-mediated deletion of UBA6 leads to a decrease in JNK 1/2 phosphorylation (Lee et al, 2011). This is intriguing as JNK1/2 is also decreased in schizophrenia (Funk et al, 2012). Among other functions, JNK1/2 is involved in phosphorylation of PSD-95 and regulation of glutamatergic transmission, which is abnormal in this illness. Our results support the idea that an ubiquitination defect may precede changes found downstream of multiple molecular pathways in schizophrenia.
The E3 ligases of UBA6 are not yet known, thus we chose to study several E3s known to be involved in synaptic transmission. We found reduced expression of Nedd4, an E3 ligase that is expressed in synapses and was recently found to mediate ubiquitination of the AMPAR (α-Amino-3-hydroxy-5-methyl-isoxazole-4-propionic acid receptor) subunit GluA1 (Lin et al, 2011; Purohit et al, 1998). At dendritic spines, DISC1 (disrupted in schizophrenia 1) interacts with TRAF2- and NCK-interacting kinase (TNIK,) promoting decreased Nedd4 activity and GluA1 ubiquitination and degradation (Brandon and Sawa, 2011). DISC1 is decreased in schizophrenia, therefore it is possible that reduced DISC1 levels could drive decreased NEDD4 in this illness, although further studies are needed to test this. In addition, Nedd4 and polyubiquitination are locally regulated in an activity-dependent manner in the dendritic spine, driving a homeostatic regulation of AMPAR in response to synaptic activity (Hou et al, 2011; Lin et al, 2011). Our results suggest a role for UPS system involvement in impairment of AMPAR homeostasis in schizophrenia.
We also found changes in polyubiquitination at specific lysines in schizophrenia. The finding that proteins of specific molecular weights are selectively affected is suggestive of specific differential mechanisms underlying abnormalities of K63 and K48 polyubiquitination in schizophrenia. This may be because of the specificity of E3 ligases. In this study, we found a decrease in K48 polyubiquitination of proteins of several molecular weights, which may suggest that a major disturbance in schizophrenia lies at the proteasome degradation level. The relative lack of protein degradation could lead to a decrease in protein turnover, resulting in less protein synthesis and the accumulation of dysfunctional, aberrant proteins. As such, schizophrenia could be considered an aggregopathy at the microcompartment level.
Although this is a feasible interpretation, the finding that K63-linked polyubiquitin is increased in schizophrenia led us consider another possibility. K63 polyubiquitination is known to have an essential role in endosomal trafficking and lysosomal degradation and, depending on the subcellular localization and specific E2s involved, E3 ligases are able to drive either K48 or K63 polyubiquitination (Sehat et al, 2008). Given that we found increased K63 polyubiquitination, it is possible that schizophrenia proteins, which are originally targeted to proteasome degradation, are diverted to lysosomes and endosomes in an attempt to maintain the intracellular balance between synthesis and degradation. It is unlikely that autophagocytosis is involved, as we found that ATG7, an E1 involved in autophagocytosis, is decreased in schizophrenia.
In addition to UPS disruption, we also found abnormalities in multiple UBL pathways. E1s involved in Nedd8ylation and URM1-MOCS3 (UBA4) were found to be decreased in schizophrenia, as were E3s involved in SUMOylation and Ufmylation. The main function of Nedd8ylation is to activate cullin Ring E3 ubiquitin ligases, which stimulate protein polyubiquitination. We found decreased in E1s and one E3 involved in this pathway in schizophrenia. These changes are likely to underlie the polyubiquitination abnormalities. The role of Nedd8ylation in neurodegenerative disorders has been characterized in Parkinson’s and Alzheimer’s disease, where Nedd8 has been shown to accumulate in inclusion bodies, neurofibrillary tangles, and senile plaques (Dil Kuazi et al, 2003; Mori et al, 2005). Given the disruption of the downstream effectors seen in schizophrenia, it is possible that a similar mechanism occurs intracellularly in this illness at a microdomain level.
Protein SUMOylation is involved in a myriad of cellular mechanisms. The redistribution of the proteins involved in SUMOylation during development is of particular interest to schizophrenia. The expression of proteins involved in SUMOylation is temporally and spatially regulated during development. Enzymes involved in SUMOylation accumulate in the nucleus during early development and redistribute to dendritic spines in adulthood (Loriol et al, 2012b). Given the key role of SUMOylation in spinogenesis (Chao et al, 2008), it is possible that delayed redistribution of SUMO components during early adulthood may lead to changes in synaptic pruning seen in schizophrenia. In addition, SUMO pathway components differentially redistribute in and out of synapses upon neuronal stimulation in an activity-dependent manner (Loriol et al, 2012a). The disruption of SUMOylation pathway proteins in schizophrenia could in turn underlie some of the characteristic features of the illness.
Ufmylation abnormalities are difficult to interpret given the lack of knowledge regarding the downstream effectors of this pathway. Recent studies have linked Ufmylation to vesicle trafficking and ER homeostasis, preventing apoptosis. The idea that ER stress response may be altered in schizophrenia was proposed in a study that showed that Sigma 1 receptors, which are protective during ER stress, are altered in schizophrenia (Mitsuda et al, 2011). Interestingly, ER stress is induced by accumulation of unfolded proteins. We show decreased expression of molecular components of the Ufmylation pathway, supporting the hypothesis that ER stress is disrupted in schizophrenia, possibly contributing to an accumulation of aberrant proteins in the ER.
In summary, our results are consistent with significant disruption of the UPS/UBL systems in schizophrenia. This study suggests novel mechanisms underlying this illness, including possible accumulation of aberrant aging proteins in cells and potential redirection of protein degradation toward the lysosomal compartment. Taken together, our findings point to a disruption in the capability of the cell to maintain homeostasis and adapt to environmental changes in schizophrenia, and provides a novel target system that may underlie aspects of the pathophysiology of this illness.
References
Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y et al (2005). Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry 58: 85–96.
Aston C, Jiang L, Sokolov BP (2004). Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res 77: 858–866.
Barta PE, Pearlson GD, Powers RE, Richards SS, Tune LE (1990). Auditory hallucinations and smaller superior temporal gyral volume in schizophrenia. Am J Psychiatry 147: 1457–1462.
Bauer D, Gupta D, Harotunian V, Meador-Woodruff JH, McCullumsmith RE (2008). Abnormal expression of glutamate transporter and transporter interacting molecules in prefrontal cortex in elderly patients with schizophrenia. Schizophr Res 104: 108–120.
Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E et al (2010a). Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet 153B: 494–502.
Bousman CA, Chana G, Glatt SJ, Chandler SD, May T, Lohr J et al (2010b). Positive symptoms of psychosis correlate with expression of ubiquitin proteasome genes in peripheral blood. Am J Med Genet B Neuropsychiatr Genet 153B: 1336–1341.
Brandon NJ, Sawa A (2011). Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci 12: 707–722.
Chao HW, Hong CJ, Huang TN, Lin YL, Hsueh YP (2008). SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J Cell Biol 182: 141–155.
Choo YS, Vogler G, Wang D, Kalvakuri S, Iliuk A, Tao WA et al (2012). Regulation of parkin and PINK1 by neddylation. Hum Mol Genet 21: 2514–2523.
Deng C, Huang XF (2006). Increased density of GABAA receptors in the superior temporal gyrus in schizophrenia. Exp Brain Res 168: 587–590.
Dil Kuazi A, Kito K, Abe Y, Shin RW, Kamitani T, Ueda N (2003). NEDD8 protein is involved in ubiquitinated inclusion bodies. J Pathol 199: 259–266.
Funk AJ, McCullumsmith RE, Haroutunian V, Meador-Woodruff JH (2012). Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacology 37: 896–905.
Geiss-Friedlander R, Melchior F (2007). Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956.
Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR et al (2000). Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry 57: 1061–1069.
Haas KF, Broadie K (2008). Roles of ubiquitination at the synapse. Biochim Biophys Acta 1779: 495–506.
Hammond JC, McCullumsmith RE, Haroutunian V, Meador-Woodruff JH (2011). Endosomal trafficking of AMPA receptors in frontal cortex of elderly patients with schizophrenia. Schizophr Res 130: 260–265.
Hou Q, Gilbert J, Man HY (2011). Homeostatic regulation of AMPA receptor trafficking and degradation by light-controlled single-synaptic activation. Neuron 72: 806–818.
Hudson JJ, Chiang SC, Wells OS, Rookyard C, El-Khamisy SF (2012). SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat Commun 3: 733.
Kadare G, Toutant M, Formstecher E, Corvol JC, Carnaud M, Boutterin MC et al (2003). PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J Biol Chem 278: 47434–47440.
Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N et al (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J 20: 4003–4012.
Kee Y, Huang M, Chang S, Moreau LA, Park E, Smith PG et al (2012). Inhibition of the Nedd8 system sensitizes cells to DNA interstrand cross-linking agents. Mol Cancer Res 10: 369–377.
Krumova P, Weishaupt JH (2012). Sumoylation in neurodegenerative diseases. Cell Mol Life Sci.
Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R et al (2000). Inhibition of the ubiquitin-proteasome system in Alzheimer’s disease. Proc Natl Acad Sci USA 97: 9902–9906.
Larimore J, Tornieri K, Ryder PV, Gokhale A, Zlatic SA, Craige B et al (2011). The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse. Mol Biol Cell 22: 4854–4867.
Lee PC, Sowa ME, Gygi SP, Harper JW (2011). Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell 43: 392–405.
Lemaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N et al (2011). Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 6: e18517.
Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F et al (2011). Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J Neurochem 119: 27–39.
Loriol C, Khayachi A, Poupon G, Gwizdek C, Martin S (2012a). Activity-dependent regulation of the sumoylation machinery in rat hippocampal neurons. Biol Cell 105: 30–45.
Loriol C, Parisot J, Poupon G, Gwizdek C, Martin S (2012b). Developmental regulation and spatiotemporal redistribution of the sumoylation machinery in the rat central nervous system. PLoS ONE 7: e33757.
Mao Y, Sun M, Desai SD, Liu LF (2000). SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci USA 97: 4046–4051.
Melchior F (2000). SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol 16: 591–626.
Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P (2002). Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci 22: 2718–2729.
Mitsuda T, Omi T, Tanimukai H, Sakagami Y, Tagami S, Okochi M et al (2011). Sigma-1Rs are upregulated via PERK/eIF2alpha/ATF4 pathway and execute protective function in ER stress. Biochem Biophys Res Commun 415: 519–525.
Moehle MS, Luduena RF, Haroutunian V, Meador-Woodruff JH, McCullumsmith RE (2012). Regional differences in expression of beta-tubulin isoforms in schizophrenia. Schizophr Res 135: 181–186.
Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H et al (2005). Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol 31: 53–61.
Nenadic I, Smesny S, Schlosser RG, Sauer H, Gaser C (2010). Auditory hallucinations and brain structure in schizophrenia: voxel-based morphometric study. Br J Psychiatry 196: 412–413.
Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM (2011). Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 14: 285–293.
Plant LD, Dowdell EJ, Dementieva IS, Marks JD, Goldstein SA (2011). SUMO modification of cell surface Kv2.1 potassium channels regulates the activity of rat hippocampal neurons. J Gen Physiol 137: 441–454.
Purohit DP, Perl DP, Haroutunian V, Powchik P, Davidson M, Davis KL (1998). Alzheimer disease and related neurodegenerative diseases in elderly patients with schizophrenia: a postmortem neuropathologic study of 100 cases. Arch Gen Psychiatry 55: 205–211.
Rajarethinam RP, DeQuardo JR, Nalepa R, Tandon R (2000). Superior temporal gyrus in schizophrenia: a volumetric magnetic resonance imaging study. Schizophr Res 41: 303–312.
Rubio MD, Haroutunian V, Meador-Woodruff JH (2012). Abnormalities of the Duo/Ras-related C3 botulinum toxin substrate 1/p21-activated kinase 1 pathway drive myosin light chain phosphorylation in frontal cortex in schizophrenia. Biol Psychiatry 71: 906–914.
Scheuring D, Kunzl F, Viotti C, San Wan Yan M, Jiang L, Schellmann S et al (2012). Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol 12: 164.
Schubert KO, Focking M, Prehn JH, Cotter DR (2012). Hypothesis review: are clathrin-mediated endocytosis and clathrin-dependent membrane and protein trafficking core pathophysiological processes in schizophrenia and bipolar disorder? Mol Psychiatry 17: 669–681.
Sehat B, Andersson S, Girnita L, Larsson O (2008). Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis. Cancer Res 68: 5669–5677.
Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R et al (2001). Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson’s disease. Science 293: 263–269.
Silbersweig DA, Stern E, Frith C, Cahill C, Holmes A, Grootoonk S et al (1995). A functional neuroanatomy of hallucinations in schizophrenia. Nature 378: 176–179.
Sun J, Maller JJ, Guo L, Fitzgerald PB (2009). Superior temporal gyrus volume change in schizophrenia: a review on region of interest volumetric studies. Brain Res Rev 61: 14–32.
Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ et al (2004). Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest 113: 1353–1363.
van der Veen AG, Ploegh HL (2012). Ubiquitin-like proteins. Annu Rev Biochem 81: 323–357.
Varshavsky A (2012). The ubiquitin system, an immense realm. Annu Rev Biochem 81: 167–176.
Vawter MP, Barrett T, Cheadle C, Sokolov BP, Wood WH 3rd, Donovan DM et al (2001). Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull 55: 641–650.
Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE (2008). Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr Res 98: 105–110.
Yin DM, Chen YJ, Sathyamurthy A, Xiong WC, Mei L (2012). Synaptic dysfunction in schizophrenia. Adv Exp Med Biol 970: 493–516.
Zheng W, Wang H, Zeng Z, Lin J, Little PJ, Srivastava LK et al (2012). The possible role of the Akt signaling pathway in schizophrenia. Brain Res 1470: 145–158.
Acknowledgements
This work was supported by National Institutes of Health Grant MH53327 (JHM-W), MH064673 (VH), and MH066392 (VH). We would like to thank Ms Jennifer Watson for her technical advice and the Alabama Brain Collection for providing material used in preliminary studies.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Rights and permissions
About this article
Cite this article
Rubio, M., Wood, K., Haroutunian, V. et al. Dysfunction of the Ubiquitin Proteasome and Ubiquitin-Like Systems in Schizophrenia. Neuropsychopharmacol 38, 1910–1920 (2013). https://doi.org/10.1038/npp.2013.84
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2013.84
Keywords
This article is cited by
-
Altered distribution and localization of organellar Na+/H+ exchangers in postmortem schizophrenia dorsolateral prefrontal cortex
Translational Psychiatry (2023)
-
Mass Spectrometry based identification of site-specific proteomic alterations and potential pathways underlying the pathophysiology of schizophrenia
Molecular Biology Reports (2023)
-
Improper Proteostasis: Can It Serve as Biomarkers for Neurodegenerative Diseases?
Molecular Neurobiology (2022)
-
Deficiency of Murine UFM1-Specific E3 Ligase Causes Microcephaly and Inflammation
Molecular Neurobiology (2022)
-
RNA-seq analysis of gene expression profiles in posttraumatic stress disorder, Parkinson’s disease and schizophrenia identifies roles for common and distinct biological pathways
Discover Mental Health (2022)