Abstract
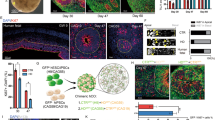
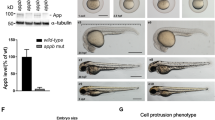
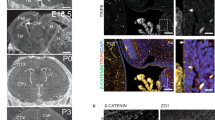
The Huntington's disease gene product, huntingtin, is indispensable for neural tube formation, but its role is obscure. We studied neurulation in htt-null embryonic stem cells and htt-morpholino zebrafish embryos and found a previously unknown, evolutionarily recent function for this ancient protein. We found that htt was essential for homotypic interactions between neuroepithelial cells; it permitted neurulation and rosette formation by regulating metalloprotease ADAM10 activity and Ncadherin cleavage. This function was embedded in the N terminus of htt and was phenocopied by treatment of htt knockdown zebrafish with an ADAM10 inhibitor. Notably, in htt-null cells, reversion of the rosetteless phenotype occurred only with expression of evolutionarily recent htt heterologues from deuterostome organisms. Conversely, all of the heterologues that we tested, including htt from Drosophila melanogaster and Dictyostelium discoideum, exhibited anti-apoptotic activity. Thus, anti-apoptosis may have been one of htt's ancestral function(s), but, in deuterostomes, htt evolved to acquire a unique regulatory activity for controlling neural adhesion via ADAM10-Ncadherin, with implications for brain evolution and development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
10 August 2012
In the version of this article initially published, the htt ATG morpholino sequence given in Online Methods (ATTTTAACAGAAGCTGTGATG) was incorrect. The correct sequence is 5′-GCCATTTTAACAGAAGCTGTGATGA-3′. The error has been corrected in the HTML and PDF versions of the article.
References
MacDonald, M.E. et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 (1993).
Tartari, M. et al. Phylogenetic comparison of huntingtin homologues reveals the appearance of a primitive polyQ in sea urchin. Mol. Biol. Evol. 25, 330–338 (2008).
Palidwor, G.A. et al. Detection of alpha-rod protein repeats using a neural network and application to huntingtin. PLoS Comput. Biol. 5, e1000304 (2009).
Myre, M.A. et al. Deficiency of huntingtin has pleiotropic effects in the social amoeba Dictyostelium discoideum. PLoS Genet. 7, e1002052 (2011).
Bhide, P.G. et al. Expression of normal and mutant huntingtin in the developing brain. J. Neurosci. 16, 5523–5535 (1996).
Zuccato, C., Valenza, M. & Cattaneo, E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol. Rev. 90, 905–981 (2010).
O'Kusky, J.R., Nasir, J., Cicchetti, F., Parent, A. & Hayden, M.R. Neuronal degeneration in the basal ganglia and loss of pallido-subthalamic synapses in mice with targeted disruption of the Huntington's disease gene. Brain Res. 818, 468–479 (1999).
Dragatsis, I., Levine, M.S. & Zeitlin, S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 26, 300–306 (2000).
Zhang, Y. et al. Depletion of wild-type huntingtin in mouse models of neurologic diseases. J. Neurochem. 87, 101–106 (2003).
Leavitt, B.R. et al. Wild-type huntingtin protects neurons from excitotoxicity. J. Neurochem. 96, 1121–1129 (2006).
Rigamonti, D. et al. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J. Neurosci. 20, 3705–3713 (2000).
Rigamonti, D. et al. Huntingtin's neuroprotective activity occurs via inhibition of procaspase-9 processing. J. Biol. Chem. 276, 14545–14548 (2001).
Ho, L.W., Brown, R., Maxwell, M., Wyttenbach, A. & Rubinsztein, D.C. Wild-type Huntingtin reduces the cellular toxicity of mutant Huntingtin in mammalian cell models of Huntington's disease. J. Med. Genet. 38, 450–452 (2001).
Duyao, M.P. et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science 269, 407–410 (1995).
Nasir, J. et al. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell 81, 811–823 (1995).
Zeitlin, S., Liu, J.P., Chapman, D.L., Papaioannou, V.E. & Efstratiadis, A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet. 11, 155–163 (1995).
Dragatsis, I., Efstratiadis, A. & Zeitlin, S. Mouse mutant embryos lacking huntingtin are rescued from lethality by wild-type extraembryonic tissues. Development 125, 1529–1539 (1998).
White, J.K. et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet. 17, 404–410 (1997).
Auerbach, W. et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum. Mol. Genet. 10, 2515–2523 (2001).
Henshall, T.L. et al. Selective neuronal requirement for huntingtin in the developing zebrafish. Hum. Mol. Genet. 18, 4830–4842 (2009).
Reiner, A. et al. Neurons lacking huntingtin differentially colonize brain and survive in chimeric mice. J. Neurosci. 21, 7608–7619 (2001).
Godin, J.D. et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron 67, 392–406 (2010).
Abranches, E. et al. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS ONE 4, e6286 (2009).
Elkabetz, Y. et al. Human ES cell–derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 22, 152–165 (2008).
Hoettecke, N., Ludwig, A., Foro, S. & Schmidt, B. Improved synthesis of ADAM10 inhibitor GI254023X. Neurodegener. Dis. 7, 232–238 (2010).
Ying, Q.L., Stavridis, M., Griffiths, D., Li, M. & Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 (2003).
Diekmann, H. et al. Decreased BDNF levels are a major contributor to the embryonic phenotype of huntingtin knockdown zebrafish. J. Neurosci. 29, 1343–1349 (2009).
Kelly, G.M. & Moon, R.T. Involvement of wnt1 and pax2 in the formation of the midbrain–hindbrain boundary in the zebrafish gastrula. Dev. Genet. 17, 129–140 (1995).
Adolf, B., Bellipanni, G., Huber, V. & Bally-Cuif, L. atoh1.2 and beta3.1 are two new bHLH-encoding genes expressed in selective precursor cells of the zebrafish anterior hindbrain. Gene Expr. Patterns 5, 35–41 (2004).
Halbleib, J.M. & Nelson, W.J. Cadherins in development: cell adhesion, sorting and tissue morphogenesis. Genes Dev. 20, 3199–3214 (2006).
Radice, G.L. et al. Developmental defects in mouse embryos lacking N-cadherin. Dev. Biol. 181, 64–78 (1997).
Reiss, K. et al. ADAM10 cleavage of N–cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signaling. EMBO J. 24, 742–752 (2005).
Marambaud, P. et al. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 21, 1948–1956 (2002).
Marcello, E. et al. Synapse-associated protein 97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J. Neurosci. 27, 1682–1691 (2007).
Malinverno, M. et al. Synaptic localization and activity of ADAM10 regulate excitatory synapses through N-cadherin cleavage. J. Neurosci. 30, 16343–16355 (2010).
Uemura, K. et al. Characterization of sequential N-cadherin cleavage by ADAM10 and PS1. Neurosci. Lett. 402, 278–283 (2006).
Lele, Z. et al. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development 129, 3281–3294 (2002).
Inoshima, I. et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 17, 1310–1314 (2011).
Chen, Y.Y., Hehr, C.L., Atkinson-Leadbeater, K., Hocking, J.C. & McFarlane, S. Targeting of retinal axons requires the metalloproteinase ADAM10. J. Neurosci. 27, 8448–8456 (2007).
Zhang, Y. et al. Huntingtin inhibits caspase-3 activation. EMBO J. 25, 5896–5906 (2006).
Yang, P., Baker, K.A. & Hagg, T. The ADAMs family: coordinators of nervous system development, plasticity and repair. Prog. Neurobiol. 79, 73–94 (2006).
Muraguchi, T. et al. RECK modulates Notch signaling during cortical neurogenesis by regulating ADAM10 activity. Nat. Neurosci. 10, 838–845 (2007).
Li, Z., Karlovich, C.A., Fish, M.P., Scott, M.P. & Myers, R.M. A putative Drosophila homolog of the Huntington's disease gene. Hum. Mol. Genet. 8, 1807–1815 (1999).
Zhang, S., Feany, M.B., Saraswati, S., Littleton, J.T. & Perrimon, N. Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington's disease model. Dis. Model Mech. 2, 247–266 (2009).
Miller, J.P. et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron 67, 199–212 (2010).
Postina, R. et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Invest. 113, 1456–1464 (2004).
Prinzen, C. et al. Differential gene expression in ADAM10 and mutant ADAM10 transgenic mice. BMC Genomics 10, 66 (2009).
Reis, S.A. et al. Striatal neurons expressing full-length mutant huntingtin exhibit decreased N-cadherin and altered neuritogenesis. Hum. Mol. Genet. 20, 2344–2355 (2011).
Westerfield, M. The Zebrafish Book (University of Oregon Press, 1993).
Thisse, C., Thisse, B., Schilling, T.F. & Postlethwait, J.H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203–1215 (1993).
Jowett, T. & Lettice, L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labeled probes. Trends Genet. 10, 73–74 (1994).
Acknowledgements
We thank M. MacDonald (Massachusetts General Hospital) for the Hdh+/+ and Hdhex4–5 cells, P. Saftig for the ADAM10 antibody (University of Kiel), E. Ruthazer for the zebrafish Ncadherin antibody (McGill University), F. Gardoni and E. Marcello for helpful technical advice on the use of ADAM10 and SAP97 antibodies, A. Badaloni for help with subcloning, and G. Simonutti for assistance with initial imaging analysis. This work has progressed at a very slow pace because it was mostly unfunded. In the beginning, this work was partially supported by the Huntington's Disease Society of America Coalition for the Cure (2007–2009) and by Italian Telethon Foundation (GGP06250, 2007–2009). Some additional support was provided by the Ministero dell'Istruzione, dell'Università e della Ricerca Scientifica, Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale (2006052993, 2006). Since 2009, progress of this work has relied entirely on occasional donations. We wish to thank one donor that in 2009 collected a considerable amount of small donations for us to continue this work and another donor that has been following our progress for 10 years. C.R. was supported by the Fundação para a Ciência e Tecnologia (grant number SFRH/BD/9627/2002) through the GABBA Programme (University of Porto). E.C., J.G. and S.Z. were members of the Huntington's Disease Society of America Coalition for the Cure and J.G. received support from NS16367.
Author information
Authors and Affiliations
Contributions
E.C., V.L.S. and C.Z. developed the study, conceived the experimental plans and analyzed the data. V.L.S. and C.Z. performed most of the biological, biochemical and molecular experiments. B.V. performed some of the biological and biochemical experiments. M.T. and C.R. participated in the initial elaboration of the project and conducted some experiments (some of the initial constructs preparation and monolayer assays, respectively). V.L.S. prepared additional constructs. M.A.M., J.A.W. and J.G. provided the Dictyostelium and Drosophila cDNA. G.G., C.Z. and A.P. performed experiments in zebrafish under the supervision of F.C. (immunocytochemistry and in situ were performed by G.G. and A.P., biochemical assays by C.Z.). M.V. and L.C. provided suggestions for some biological experiments. S.Z. provided the conditional knockout mice and some constructs. B.D. and B.S. provided GI254023X. V.L.S., C.Z. and E.C. interpreted the data and wrote the manuscript. All of the authors read and edited the manuscript. E.C. supervised the entire work, directed the strategies, provided financial support and gave final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 19081 kb)
Supplementary Video 1
Time lapse Imaging experiment in Hdh+/+ cells during neural differentiation. (MOV 14150 kb)
Supplementary Video 2
Time lapse Imaging experiment in Hdhex4/5 cells during neural differentiation. (MOV 14455 kb)
Supplementary Video 3
Time lapse Imaging experiment of co-culture system during neural differentiation. (MOV 21124 kb)
Rights and permissions
About this article
Cite this article
Lo Sardo, V., Zuccato, C., Gaudenzi, G. et al. An evolutionary recent neuroepithelial cell adhesion function of huntingtin implicates ADAM10-Ncadherin. Nat Neurosci 15, 713–721 (2012). https://doi.org/10.1038/nn.3080
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3080
This article is cited by
-
MAP4K4 promotes ovarian cancer metastasis through diminishing ADAM10-dependent N-cadherin cleavage
Oncogene (2023)
-
Contribution of Glial Cells to Polyglutamine Diseases: Observations from Patients and Mouse Models
Neurotherapeutics (2023)
-
Genome-wide screening in pluripotent cells identifies Mtf1 as a suppressor of mutant huntingtin toxicity
Nature Communications (2023)
-
The evolutionary history of the polyQ tract in huntingtin sheds light on its functional pro-neural activities
Cell Death & Differentiation (2022)
-
RNA-seq analysis reveals significant transcriptome changes in huntingtin-null human neuroblastoma cells
BMC Medical Genomics (2021)