Abstract
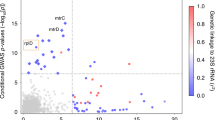
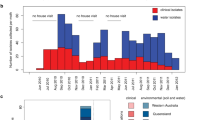
Bacteria pose unique challenges for genome-wide association studies because of strong structuring into distinct strains and substantial linkage disequilibrium across the genome1,2. Although methods developed for human studies can correct for strain structure3,4, this risks considerable loss-of-power because genetic differences between strains often contribute substantial phenotypic variability5. Here, we propose a new method that captures lineage-level associations even when locus-specific associations cannot be fine-mapped. We demonstrate its ability to detect genes and genetic variants underlying resistance to 17 antimicrobials in 3,144 isolates from four taxonomically diverse clonal and recombining bacteria: Mycobacterium tuberculosis, Staphylococcus aureus, Escherichia coli and Klebsiella pneumoniae. Strong selection, recombination and penetrance confer high power to recover known antimicrobial resistance mechanisms and reveal a candidate association between the outer membrane porin nmpC and cefazolin resistance in E. coli. Hence, our method pinpoints locus-specific effects where possible and boosts power by detecting lineage-level differences when fine-mapping is intractable.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Feil, E. J. & Spratt, B. G. Recombination and the structures of bacterial pathogens. Annu. Rev. Microbiol. 55, 561–590 (2001).
Falush, D. & Bowden, R. Genome-wide association mapping in bacteria? Trends Microbiol. 14, 353–355 (2006).
Stephens, M. & Balding, D. J. Bayesian statistical methods for genetic association studies. Nature Rev. Genet. 10, 681–690 (2009).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Cordero, O. X. & Polz, M. F. Explaining microbial genomic diversity in light of evolutionary ecology. Nature Rev. Microbiol. 12, 263–273 (2014).
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998).
Falkowski, P. G., Fenchel, T. & Delong, E. F. The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039 (2008).
World Health Organization. The Global Burden of Disease: 2004 Update (2008); http://www.who.int/healthinfo/global_burden_disease
Davies, J. & Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 (2010).
European Centre for Disease Prevention and Control. Surveillance of Surgical-Site Infections in Europe, 2008–2009 (2012); http://www.ecdc.europa.eu/en/publications/Publications/120215_SUR_SSI_2008-2009.pdf
World Health Organization. Global Tuberculosis Report 2014 (2014); http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf
World Health Organization. Antimicrobial Resistance: A Global Report on Surveillance (2014); http://www.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf
Sheppard, S. K. et al. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc. Natl Acad. Sci. USA 110, 11923–11927 (2013).
Alam, M. T. et al. Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol. Evol. 6, 1174–1185 (2014).
Laabei, M. et al. Predicting the virulence of MRSA from its genome sequence. Genome Res. 24, 839–849 (2014).
Chewapreecha, C. et al. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet. 10, e1004547 (2014).
Salipante, S. J. et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res. 25, 119–128 (2014).
Read, T. D. & Massey, R. C. Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: a new direction for bacteriology. Genome Med. 6, 109 (2014).
Fahrat, M. R., Shapiro, B. J., Sheppard, S. K., Colijn, C. & Murray, M. A phylogeny-based sampling strategy and power calculator informs genome-wide associations study design for microbial pathogens. Genome Med. 6, 101 (2014).
Hall, B. G. SNP-associations and phenotype predictions from hundreds of microbial genomes without genome alignments. PLoS ONE 9, e90490 (2014).
Chen, P. E. & Shapiro, B. J. The advent of genome-wide association studies for bacteria. Curr. Opin. Microbiol. 25, 17–24 (2015).
Holt, K. E. et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl Acad. Sci. USA 112, E3574–E3581 (2015).
Price, A. L., Zaitlen, N. A., Reich, D. & Patterson, N. New approaches to population stratification in genome-wide association studies. Nature Rev. Genet. 11, 459–463 (2010).
Perez-Losada, M. et al. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect. Genet. Evol. 6, 97–112 (2006).
Vos, M. & Didelot, X. A comparison of homologous recombination rates in bacteria and archeae. IMSE J. 3, 199–208 (2009).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 38, 904–909 (2006).
O'Neill, A. J., McLaws, F., Kahlmeter, G., Henriksen, A. S. & Chopra, I. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob. Agents Chemother. 51, 1737–1740 (2007).
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nature Genet. 44, 821–824 (2012).
Yang, J., Zaitlen, N. A., Goddard, M. E., Visscher, P. M. & Price, A. L. Advantages and pitfalls in the application of mixed-model association methods. Nature Genet. 46, 100–106 (2014).
Grafen, A. The phylogenetic regression. Phil. Trans. R. Soc. Lond. B 326, 119–157 (1989).
Martins, E. P. & Hansen, T. F. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667 (1997).
Milkman, R. & Bridges, M. M. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics 126, 505–517 (1990).
McVean, G. A genealogical interpretation of principal components analysis. PLoS Genet. 5, e1000686 (2009).
Astle, W. & Balding, D. J. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 24, 451–471 (2009).
Wald, A. Tests of statistical hypotheses concerning several parameters when the number of observations is large. Trans. Am. Math. Soc. 54, 426–482 (1943).
Walker, T. M. et al. Whole genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect. Dis. 15, 1193–1202 (2015).
Gordon, N. C. et al. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J. Clin. Microbiol. 52, 1182–1191 (2014).
Stoesser, N. et al. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genome sequence data. J. Antimicrob. Chemother. 68, 2234–2244 (2013).
Bradley, P. et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nature Commun. 6, 10063 (2015).
Sun, S., Berg, O. G., Roth, J. R. & Andersson, D. I. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182, 1183–1195 (2009).
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genet. 38, 203–208 (2006).
Kang, H. M. et al. Efficient control of population structure in model organism association mapping. Genetics 178, 1709–1723 (2008).
Kang, H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nature Genet. 42, 348–354 (2010).
Lippert, C. et al. FaST linear mixed models for genome-wide association studies. Nature Methods 8, 833–835 (2011).
Listgarten, J. et al. Improved linear mixed models for genome-wide association studies. Nature Methods 9, 525–526 (2012).
O'Hagan, A. & Forster, J. in Kendall's Advanced Theory of Statistics Volume 2B Bayesian Inference 2nd edn, Ch. 11 (Wiley-Blackwell, 2010).
Eyre, D. W. et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2, e001124 (2012).
Everitt, R. G. et al. Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nature Commun. 5, 3956 (2014).
Lunter, G. & Goodson, M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21, 936–939 (2011).
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Rizk, G., Lavenier, D. & Chikhi, R. DSK: k-mer counting with very low memory usage. Bioinformatics 29, 652–653 (2013).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing data inference for whole genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Didelot, X. & Wilson, D. J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 11, e1004041 (2015).
Hedge, J. & Wilson, D. J. Bacterial phylogenetic reconstruction from whole genomes is robust to recombination but demographic inference is not. mBio 5, e02158–14 (2014).
Pupko, T., Pe'er, I., Shamir, R. & Graur, D. A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol. Biol. Evol. 17, 890–896 (2000).
Yahara, K., Didelot, X., Ansari, M., Sheppard, S. K. & Falush, D. Efficient inference of recombination hot regions in bacterial genomes. Mol. Biol. Evol. 31, 1593–1605 (2014).
Dunn, O. J. Estimation of the medians for dependent variables. Ann. Math. Stat. 30, 192–197 (1959).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 431 (2009).
Langmead, B. & Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012).
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Acknowledgements
The authors thank J.-B. Veyrieras, D. Charlesworth and B. Charlesworth for comments on the manuscript, X. Zhou and M. Stephens for helping adapt their software, S. Niemann for assisting with tuberculosis isolates and X. Didelot, D. Falush, R. Bowden, S. Myers, J. Marchini, J. Pickrell, P. Visscher, A. Price and P. Donnelly for discussions. This study was supported by the Oxford NIHR Biomedical Research Centre, a Mérieux Research Grant and the UKCRC Modernising Medical Microbiology Consortium, the latter funded under the UKCRC Translational Infection Research Initiative supported by the Medical Research Council, the Biotechnology and Biological Sciences Research Council and the National Institute for Health Research on behalf of the UK Department of Health (grant no. G0800778) and the Wellcome Trust (grant no. 087646/Z/08/Z). T.M.W. is an MRC research training fellow. C.C.A.S. was supported by a Wellcome Trust Career Development Fellowship (grant no. 097364/Z/11/Z). D.A.C. is funded by the Royal Academy of Engineering and an EPSRC Healthcare Technologies Challenge Award. T.E.P. and D.W.C. are NIHR Senior Investigators. G.M. is supported by a Wellcome Trust Investigator Award (grant no. 100956/Z/13/Z). D.J.W. and Z.I. are Sir Henry Dale Fellows, jointly funded by the Wellcome Trust and the Royal Society (grants nos. 101237/Z/13/Z and 102541/Z/13/Z).
Author information
Authors and Affiliations
Contributions
S.G.E., C.-H.W., J.C. and D.J.W. designed the study, developed the methods, performed the analysis, interpreted the results and wrote the manuscript. Z.I. and D.A.C. assisted the analysis and commented on the manuscript. N.S., N.C.G., T.M.W., K.L.H., N.W., E.G.S., N.I., M.J.L., T.E.P. and D.W.C. designed and implemented isolate collection, drug susceptibility testing and whole-genome sequencing, and assisted with interpretation. C.C.A.S., G.M. and A.S.W. assisted with methods development and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1-10 and Tables 1-5 (PDF 18390 kb)
Supplementary Data 1
Individual BioSample accession numbers and antimicrobial resistance phenotypes. (XLSX 260 kb)
Rights and permissions
About this article
Cite this article
Earle, S., Wu, CH., Charlesworth, J. et al. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol 1, 16041 (2016). https://doi.org/10.1038/nmicrobiol.2016.41
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.41
This article is cited by
-
happi: a hierarchical approach to pangenomics inference
Genome Biology (2023)
-
Detecting patterns of accessory genome coevolution in Staphylococcus aureus using data from thousands of genomes
BMC Bioinformatics (2023)
-
Laboratory diagnosed microbial infection in English UK Biobank participants in comparison to the general population
Scientific Reports (2023)
-
Gut microbial structural variation associates with immune checkpoint inhibitor response
Nature Communications (2023)
-
Genomic insights into zoonotic transmission and antimicrobial resistance in Campylobacter jejuni from farm to fork: a one health perspective
Gut Pathogens (2022)