Abstract
While recent studies have identified higher than anticipated heterogeneity of mutation rate across genomic regions, mutations in exons and introns are assumed to be generated at the same rate. Here we find fewer somatic mutations in exons than expected from their sequence content and demonstrate that this is not due to purifying selection. Instead, we show that it is caused by higher mismatch-repair activity in exonic than in intronic regions. Our findings have important implications for understanding of mutational and DNA repair processes and knowledge of the evolution of eukaryotic genes, and they have practical ramifications for the study of evolution of both tumors and species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chapman, M.A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Dulak, A.M. et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 45, 478–486 (2013).
Pleasance, E.D. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2010).
Li, J. et al. A dual model for prioritizing cancer mutations in the non-coding genome based on germline and somatic events. PLoS Comput. Biol. 11, e1004583 (2015).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Koren, A. et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am. J. Hum. Genet. 91, 1033–1040 (2012).
Stamatoyannopoulos, J.A. et al. Human mutation rate associated with DNA replication timing. Nat. Genet. 41, 393–395 (2009).
Hodgkinson, A. & Eyre-Walker, A. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 12, 756–766 (2011).
Polak, P. et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 (2015).
Schuster-Böckler, B. & Lehner, B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504–507 (2012).
Morganella, S. et al. The topography of mutational processes in breast cancer genomes. Nat. Commun. 7, 11383 (2016).
Perera, D. et al. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature 532, 259–263 (2016).
Polak, P. et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat. Biotechnol. 32, 71–75 (2014).
Sabarinathan, R., Mularoni, L., Deu-Pons, J., Gonzalez-Perez, A. & López-Bigas, N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature 532, 264–267 (2016).
Li, F. et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 153, 590–600 (2013).
Tatum, D. & Li, S. Evidence that the histone methyltransferase Dot1 mediates global genomic repair by methylating histone H3 on lysine 79. J. Biol. Chem. 286, 17530–17535 (2011).
House, N.C.M., Koch, M.R. & Freudenreich, C.H. Chromatin modifications and DNA repair: beyond double-strand breaks. Front. Genet. 5, 296 (2014).
Schwartz, S., Meshorer, E. & Ast, G. Chromatin organization marks exon–intron structure. Nat. Struct. Mol. Biol. 16, 990–995 (2009).
Huff, J.T., Plocik, A.M., Guthrie, C. & Yamamoto, K.R. Reciprocal intronic and exonic histone modification regions in humans. Nat. Struct. Mol. Biol. 17, 1495–1499 (2010).
Fredriksson, N.J., Ny, L., Nilsson, J.A. & Larsson, E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 46, 1258–1263 (2014).
Hodis, E. et al. A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012).
Lanzós, A. et al. Discovery of cancer driver long noncoding RNAs across 1112 tumour genomes: new candidates and distinguishing features. Sci. Rep. 7, 41544 (2017).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Futreal, P.A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Rubio-Perez, C. et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27, 382–396 (2015).
Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 (2008).
Hause, R.J., Pritchard, C.C., Shendure, J. & Salipante, S.J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 22, 1342–1350 (2016).
Shlien, A. et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 47, 257–262 (2015).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Marteijn, J.A., Lans, H., Vermeulen, W. & Hoeijmakers, J.H.J. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 (2014).
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Adar, S., Hu, J., Lieb, J.D. & Sancar, A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc. Natl. Acad. Sci. USA 113, E2124–E2133 (2016).
Hu, J., Adar, S., Selby, C.P., Lieb, J.D. & Sancar, A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 (2015).
Haradhvala, N.J. et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 164, 538–549 (2016).
Tolstorukov, M.Y., Volfovsky, N., Stephens, R.M. & Park, P.J. Impact of chromatin structure on sequence variability in the human genome. Nat. Struct. Mol. Biol. 18, 510–515 (2011).
Francioli, L.C. et al. Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 47, 822–826 (2015).
Supek, F. & Lehner, B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 521, 81–84 (2015).
Supek, F. & Lehner, B. Clustered mutation signatures reveal that error-prone DNA repair targets mutations to active genes. Cell 170, 534–547 (2017).
Kim, S., Kim, H., Fong, N., Erickson, B. & Bentley, D.L. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc. Natl. Acad. Sci. USA 108, 13564–13569 (2011).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell http://dx.doi.org/10.1016/j.cell.2017.09.042 (2017).
Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 107, 961–968 (2010).
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41, 376–381 (2009).
Chamary, J.V., Parmley, J.L. & Hurst, L.D. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7, 98–108 (2006).
Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005).
Hoffman, M.M. & Birney, E. Estimating the neutral rate of nucleotide substitution using introns. Mol. Biol. Evol. 24, 522–531 (2007).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Acknowledgements
We acknowledge funding from the Spanish Ministry of Economy and Competitiveness (SAF2015-66084-R, MINECO/FEDER, UE), La Fundació la Marató de TV3, EU H2020 Programme 2014-2020 under grant agreements 634143 (MedBioinformatics) and by the European Research Council (Consolidator Grant 682398). IRB Barcelona is the recipient of a Severo Ochoa Centre of Excellence Award from the Spanish Ministry of Economy and Competitiveness (MINECO; Government of Spain) and is supported by CERCA (Generalitat de Catalunya). R.S. is supported by an EMBO Long-Term Fellowship (ALTF 568-2014) cofunded by the European Commission (EMBOCOFUND2012, GA-2012-600394) with support from Marie Curie Actions. A.G.-P. is supported by a Ramón y Cajal contract from the Spanish Ministry of Economy and Competitiveness (RYC-2013-14554). We acknowledge the contribution of I. Reyes-Salazar to refactoring and cleaning all code produced in the study for publication. We are grateful to B. Campbell and U. Tabori for help in obtaining the mutation calls for bMMRD samples sequenced by the International BMMRD Consortium. The results published here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
Author information
Authors and Affiliations
Contributions
J.F. and R.S. participated in the design and execution of analyses, produced the figures, participated in the interpretation of results and edited the manuscript. L.M. developed computational code employed in the analyses. F.M. developed the statistical framework to compute the significance of the decreased exonic mutation burden and its correlation with chromatin features. A.G.-P. participated in the design of analyses, the interpretation of results, the oversight of analyses, and drafted and edited the manuscript. N.L.-B. conceived the study, participated in the design of analyses, oversaw the study and the interpretation of results, and drafted and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Relationship between the exon/intron proportion of genes and their mutation rate.
Each box plot represents the distribution of mutation rate (in colorectal POLE-mutant tumors) of genes in deciles of increasing exon to intron proportion. This relationship implies that computing a mutation rate in exons and introns by aggregating genes with different mutation rates and exon to intron proportions would create an artifact. To avoid this artifact, we compute the expected mutations in exons and introns for each gene (Fig. 2).
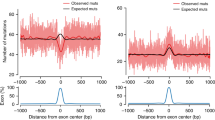
Supplementary Figure 2 Permutation-computed expected mutation rate.
(a) Exon-centered plot of observed and expected mutation rates for several clusters of tumors: top row, colorectal and uterine POLE-mutant tumors; top and bottom rows, colorectal and uterine MSI-H tumors; bottom row, bMMRD POLE- and POLD-mutant tumors (analogous to Figs. 2a and 3a). The expected mutation rate was computed using 1,000 random permutations of mutations in each sequence of the stack, as described in the Online Methods. (b) Density plot representing the difference between the observed and expected number of exonic mutations in individual genes with at least one expected exonic mutation (similar to Fig. 2c).
Supplementary Figure 3 Decreased exonic mutation burden across groups of genes with increasing mutation rate.
Graphs to the left present the decreased exonic mutation burden of groups of genes of increasing mutation rate (circles in the graphs). In each graph, groups of genes with significantly decreased exonic mutation burden are colored in purple, while those with non-significantly decreased exonic mutation burden are colored in blue. Graphs to the right present the distribution of mutation rates for each group of genes.
Supplementary Figure 4 Relationship between decreased exonic mutation burden and exonic enrichment of H3K36me3 (read count based) across clusters of tumors.
From top to bottom, the rows represent the relationship between exonic enrichment of H3K36me3 (read count based as explained in the Online Methods) and decreased exonic mutation burden in colorectal POLE-mutant tumors, colorectal MSI-H tumors, bMMRD POLE-mutant tumors and bMMRD POLD-mutant tumors. From left to right, the correlations have been computed grouping the genes into 10, 25 and 50 bins (similar to Fig. 4).
Supplementary Figure 5 Relationship between decreased exonic mutation burden and exonic enrichment of H3K36me3 (peak based) across clusters of tumors.
From top to bottom, the rows represent the relationship between exonic enrichment of H3K36me3 and decreased exonic mutation burden in colorectal POLE-mutant tumors, colorectal MSI-H tumors, bMMRD POLE-mutant tumors and bMMRD POLD-mutant tumors. The exonic enrichment for H3K36me3 (exon to intron ratio) has been calculated from H3K36me3 peaks detected in the closest representative in Roadmap Epigenomics to the cell of origin of each tumor. From left to right, the correlations have been computed grouping the genes into 10, 25 and 50 bins (similar to Fig. 4). We corroborate that the same negative correlation is obtained between exonic enrichment for H3K36me3 and decreased exonic mutation rate if peaks instead of ChIP–seq reads (as in Fig. 4) are used to compute the former. The three panels at the bottom represent the correlation between the exon to intron ratio of nucleosomes (computed from nucleosome occupancy; Online Methods) and the decreased exonic mutation rate in POLE-mutant tumors.
Supplementary Figure 6 Clustering of tumors of different cancer types according to their mutational signatures.
The approach to build the clusters is described in the Online Methods.
Supplementary Figure 7 Exon-centered nucleotide-excision repair efficiency.
(a) The first row shows the exon-centered frequency of dipyrimidines (TT, TC, CT and CC, from left to right) favorable for UV-induced damage. The second row represents the frequency of dipyrimidines that are covered by at least one XR–seq read (with matching dipyrimdine at positions 19–20 of the reads) for CPD repair in NHF1 cell lines. The last row represents the ratio of the two previous rows. (b) Quantification of the ratio computed above for exonic and intronic dipyrimidines across three cell lines with different NER pathways at play, both for CPDs and 6,4 photoproducts (see Online Methods for details).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1562 kb)
Supplementary Table 1
Coverage of several chromatin features of exons and introns across the structure of genes. (XLSX 1368 kb)
Supplementary Table 2
Difference in exonic and intronic coverage (Mann–Whitney P value) and mean exonic coverage across the genic structure. (XLSX 222 kb)
Supplementary Table 3
Decreased exonic mutation rate across clusters of tumors. (XLSX 10 kb)
Supplementary Table 4
Decreased exonic mutation burden across groups of genes with different mutation rate covariate values. (XLSX 38 kb)
Supplementary Table 5
Decreased exonic mutation burden for individual tumors. (XLSX 55 kb)
Supplementary Table 6
Correlation of decreased exonic mutation burden and the exonic enrichment for several histone marks. (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Frigola, J., Sabarinathan, R., Mularoni, L. et al. Reduced mutation rate in exons due to differential mismatch repair. Nat Genet 49, 1684–1692 (2017). https://doi.org/10.1038/ng.3991
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3991
This article is cited by
-
Non-canonical antigens are the largest fraction of peptides presented by MHC class I in mismatch repair deficient murine colorectal cancer
Genome Medicine (2024)
-
The need for assessment of risks arising from interactions between NGT organisms from an EU perspective
Environmental Sciences Europe (2023)
-
Mutation bias reflects natural selection in Arabidopsis thaliana
Nature (2022)
-
Complex genomic patterns of abasic sites in mammalian DNA revealed by a high-resolution SSiNGLe-AP method
Nature Communications (2022)
-
The shaping of cancer genomes with the regional impact of mutation processes
Experimental & Molecular Medicine (2022)