Abstract
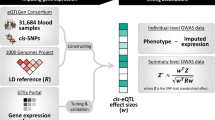
Genetic association mapping produces statistical links between phenotypes and genomic regions, but identifying causal variants remains difficult. Whole-genome sequencing (WGS) can help by providing complete knowledge of all genetic variants, but it is financially prohibitive for well-powered GWAS studies. We performed mapping of expression quantitative trait loci (eQTLs) with WGS and RNA-seq, and found that lead eQTL variants called with WGS were more likely to be causal. Through simulations, we derived properties of causal variants and used them to develop a method for identifying likely causal SNPs. We estimated that 25–70% of causal variants were located in open-chromatin regions, depending on the tissue and experiment. Finally, we identified a set of high-confidence causal variants and showed that these were more enriched in GWAS associations than other eQTLs. Of those, we found 65 associations with GWAS traits and provide examples in which genes implicated by expression are functionally validated as being relevant for complex traits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Spain, S.L. & Barrett, J.C. Strategies for fine-mapping complex traits. Hum. Mol. Genet. 24, R111–R119 (2015).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
Brown, A.A. et al. Genetic interactions affecting human gene expression identified by variance association mapping. eLife 3, e01381 (2014).
Buil, A. et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet. 47, 88–91 (2015).
UK10K Consortium. et al. The UK10K project identifies rare variants in health and disease. Nature 526, 82–90 (2015).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Timpson, N.J. et al. A rare variant in APOC3 is associated with plasma triglyceride and VLDL levels in Europeans. Nat. Commun. 5, 4871 (2014).
Iotchkova, V. et al. Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet. 48, 1303–1312 (2016).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Lebreton, C.M. & Visscher, P.M. Empirical nonparametric bootstrap strategies in quantitative trait loci mapping: conditioning on the genetic model. Genetics 148, 525–535 (1998).
Visscher, P.M., Thompson, R. & Haley, C.S. Confidence intervals in QTL mapping by bootstrapping. Genetics 143, 1013–1020 (1996).
Hormozdiari, F., Kostem, E., Kang, E.Y., Pasaniuc, B. & Eskin, E. Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014).
Wen, X., Lee, Y., Luca, F. & Pique-Regi, R. Efficient integrative multi-SNP association analysis via deterministic approximation of posteriors. Am. J. Hum. Genet. 98, 1114–1129 (2016).
Manolio, T.A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Nica, A.C. et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 6, e1000895 (2010).
Ongen, H. et al. Estimating the causal tissues for complex traits and diseases. Nat. Genet. http://dx.doi.org/10.1038/ng.3981 2017).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Sharma, G. et al. GPER deficiency in male mice results in insulin resistance, dyslipidemia, and a proinflammatory state. Endocrinology 154, 4136–4145 (2013).
Meyre, D. et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 41, 157–159 (2009).
Jelinek, D., Heidenreich, R.A., Erickson, R.P. & Garver, W.S. Decreased Npc1 gene dosage in mice is associated with weight gain. Obesity (Silver Spring) 18, 1457–1459 (2010).
Jelinek, D. et al. Npc1 haploinsufficiency promotes weight gain and metabolic features associated with insulin resistance. Hum. Mol. Genet. 20, 312–321 (2011).
Bambace, C., Dahlman, I., Arner, P. & Kulyté, A. NPC1 in human white adipose tissue and obesity. BMC Endocr. Disord. 13, 5 (2013).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
Hormozdiari, F. et al. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99, 1245–1260 (2016).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Marco-Sola, S., Sammeth, M., Guigó, R. & Ribeca, P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat. Methods 9, 1185–1188 (2012).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Ongen, H., Buil, A., Brown, A.A., Dermitzakis, E.T. & Delaneau, O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485 (2016).
Flutre, T., Wen, X., Pritchard, J. & Stephens, M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 9, e1003486 (2013).
Wen, X., Luca, F. & Pique-Regi, R. Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS Genet. 11, e1005176 (2015).
Servin, B. & Stephens, M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3, e114 (2007).
The International Multiple Sclerosis Genetics Consortium. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45, 1353–1360 (2013).
Chen, W. et al. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics 200, 719–736 (2015).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Robinson, E.B. et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet. 48, 552–555 (2016).
Horikoshi, M. et al. Genome-wide associations for birth weight and correlations with adult disease. Nature 538, 248–252 (2016).
Locke, A.E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Nikpay, M. et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Liu, J.Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Fuchsberger, C. et al. The genetic architecture of type 2 diabetes. Nature 536, 41–47 (2016).
Manning, A.K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44, 659–669 (2012).
Wood, A.R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Willer, C.J. et al.; Global Lipid Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Delaneau, O. et al. A complete tool set for molecular QTL discovery and analysis. Nat. Commun. 8, 15452 (2017).
Acknowledgements
We thank N. Lykoskoufis for assistance with the enrichment analysis. T.S. is supported as an NIHR Senior Research Fellow. This project was supported by a Helse Sør-Øst grant (2011060) to A.B. and an MRC Project Grant (L01999X/1) to K.S., and by grants from the NIH-NIMH (NIH-R01MH101814-GTEx), an IMI-Joint Undertaking of the European Commission (UE7-DIRECT-115317-1), the European Commission (UE7-EUROBATS-259749), the European Research Council (UE7-POPRNASEQ-260927), the Louis Jeantet Foundation, the Swiss National Science Foundation (31003A-149984 and 31003A-170096), and SystemsX (2012/201-SysGenetix) to E.T.D. The TwinsUK study was funded by the Wellcome Trust; European Community's Seventh Framework Programme (FP7/2007-2013) and the Medical Research Council. The study also received support from the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre, based at Guy's and St Thomas' NHS Foundation Trust, in partnership with King's College London. SNP genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH-CIDR. This study used data generated by the UK10K Consortium. Funding for UK10K was provided by the Wellcome Trust under award WT091310. A full list of the investigators who contributed to the generation of the UK10K data is available at http://www.UK10K.org/. This research was supported by grants from the European Research Council. Computation was performed at the Vital-IT Center (http://www.vital-it.ch/) for high-performance computing of the SIB Swiss Institute of Bioinformatics.
Author information
Authors and Affiliations
Contributions
A.A.B. and E.T.D. designed the study. A.A.B. ran the analyses. A.A.B., A.V., and E.T.D. interpreted the results. A.A.B., A.V., and E.T.D. wrote the manuscript. O.D. provided methodological suggestions. K.S.S. and T.D.S. contributed data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Rank of statistical association for the causal variant in simulations.
Based on five simulations per tissue, the x-axis shows the rank of the causal variant, and the y-axis the proportion of times this outcome occurred. We notice that, as the whole blood experiment was smaller than the other experiments, sample size does not seem to affect the distribution. The causal variant is the most associated variant in 45% of cases, and among the ten most significantly associated variants 89% of times. The boxes show the 25th and 75th percentiles, and the whiskers end at the furthest value from the edge of the box that is not more than 1.5 times the inter-quartile range. Any values outside these whiskers are outliers that are plotted directly.
Supplementary Figure 2 Minor allele frequencies of LEVs called with the two technologies.
The LEVs called using sequence have a lower minor allele frequency than those called using arrays (0.26 vs. 0.27). The box edges show the 25th and 75th percentiles, and the whiskers end at the maximum and minimum values.
Supplementary Figure 3 Impute-derived INFO scores for the two genotyping arrays of the sequence LEVs.
The dots in pink reflect LEVs that were filtered from the array data due to poor imputation quality.
Supplementary Figure 4 Relationship between CaVEMaN score and causal probability in simulations.
The CaVEMaN score is calibrated using the simulations to estimate the probability that the lead eQTL variant is causal. The estimated calibration functions are consistent across tissues, with the exception of blood, which is slightly less conservative than the other tissues, probably due to the smaller sample size.
Supplementary Figure 5 Validation of CaVEMaN probabilities with the Geuvadis dataset.
Five simulated datasets were produced based on the genotype data and eQTLs mapped in Geuvadis. CaVEMaN was run on all of these datasets, and we plot the median CaVEMaN causal probabilities for LEVs, binned into 10 groups, against the true proportions of LEVs in these bins that were causal on the trait. We also show on this plot the equivalent analysis performed using dap-g and a further simulation where the assumption of only one genetic signal in the region is violated by simulating a secondary eQTL.
Supplementary Figure 6 Comparison between CaVEMaN and CAVIAR estimates of causal probabilities.
The comparison was only performed for genes with only one eQTL to minimize differences due to multiple eQTL mapping strategies. Spearman correlation between the two estimates was 0.856.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1 and 2 and Supplementary Note
Supplementary Data Set 1
A full list of all eQTLs discovered in the five experiments, together with P value for association and causal probability score
Supplementary Data Set 2
A list of high confidence causal variants which are also significantly associated with a GWAS trait, together with an estimate produced by coloc of the probability of a shared genetic signal
Rights and permissions
About this article
Cite this article
Brown, A., Viñuela, A., Delaneau, O. et al. Predicting causal variants affecting expression by using whole-genome sequencing and RNA-seq from multiple human tissues. Nat Genet 49, 1747–1751 (2017). https://doi.org/10.1038/ng.3979
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3979
This article is cited by
-
Identifying novel regulatory effects for clinically relevant genes through the study of the Greek population
BMC Genomics (2023)
-
Molecular quantitative trait loci
Nature Reviews Methods Primers (2023)
-
Integrative genomic analyses of promoter G-quadruplexes reveal their selective constraint and association with gene activation
Communications Biology (2023)
-
DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits
Nature Genetics (2023)
-
Genetic analysis of blood molecular phenotypes reveals common properties in the regulatory networks affecting complex traits
Nature Communications (2023)