Abstract
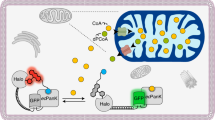
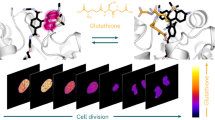
Methionine can be reversibly oxidized to methionine sulfoxide (MetO) under physiological and pathophysiological conditions, but its use as a redox marker suffers from the lack of tools to detect and quantify MetO within cells. In this work, we created a pair of complementary stereospecific genetically encoded mechanism-based ratiometric fluorescent sensors of MetO by inserting a circularly permuted yellow fluorescent protein between yeast methionine sulfoxide reductases and thioredoxins. The two sensors, respectively named MetSOx and MetROx for their ability to detect S and R forms of MetO, were used for targeted analysis of protein oxidation, regulation and repair as well as for monitoring MetO in bacterial and mammalian cells, analyzing compartment-specific changes in MetO and examining responses to physiological stimuli.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
D'Autréaux, B. & Toledano, M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 (2007).
Finkel, T. Signal transduction by reactive oxygen species. J. Cell Biol. 194, 7–15 (2011).
Meyer, A.J. & Dick, T.P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 13, 621–650 (2010).
Maron, B.A. & Michel, T. Subcellular localization of oxidants and redox modulation of endothelial nitric oxide synthase. Circ. J. 76, 2497–2512 (2012).
Nagai, T., Sawano, A., Park, E.S. & Miyawaki, A. Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc. Natl. Acad. Sci. USA 98, 3197–3202 (2001).
Belousov, V.V. et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 3, 281–286 (2006).
Malinouski, M., Zhou, Y., Belousov, V.V., Hatfield, D.L. & Gladyshev, V.N. Hydrogen peroxide probes directed to different cellular compartments. PLoS ONE 6, e14564 (2011).
Niethammer, P., Grabher, C., Look, A.T. & Mitchison, T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459, 996–999 (2009).
Jin, B.Y., Sartoretto, J.L., Gladyshev, V.N. & Michel, T. Endothelial nitric oxide synthase negatively regulates hydrogen peroxide–stimulated AMP-activated protein kinase in endothelial cells. Proc. Natl. Acad. Sci. USA 106, 17343–17348 (2009).
Østergaard, H., Henriksen, A., Hansen, F.G. & Winther, J.R. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 20, 5853–5862 (2001).
Hanson, G.T. et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279, 13044–13053 (2004).
Morgan, B. et al. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 9, 119–125 (2013).
Le, D.T. et al. Analysis of methionine/selenomethionine oxidation and methionine sulfoxide reductase function using methionine-rich proteins and antibodies against their oxidized forms. Biochemistry 47, 6685–6694 (2008).
Wehr, N.B. & Levine, R.L. Wanted and wanting: antibody against methionine sulfoxide. Free Radic. Biol. Med. 53, 1222–1225 (2012).
Brot, N., Weissbach, L., Werth, J. & Weissbach, H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc. Natl. Acad. Sci. USA 78, 2155–2158 (1981).
Grimaud, R. et al. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276, 48915–48920 (2001).
Kryukov, G.V., Kumar, R.A., Koc, A., Sun, Z. & Gladyshev, V.N. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 99, 4245–4250 (2002).
Tarrago, L. & Gladyshev, V.N. Recharging oxidative protein repair: catalysis by methionine sulfoxide reductases towards their amino acid, protein, and model substrates. Biochemistry (Mosc.) 77, 1097–1107 (2012).
Boschi-Muller, S., Olry, A., Antoine, M. & Branlant, G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim. Biophys. Acta 1703, 231–238 (2005).
Motohashi, K., Kondoh, A., Stumpp, M.T. & Hisabori, T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. USA 98, 11224–11229 (2001).
Zhao, Y. et al. An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891 (2011).
Bilan, D.S. et al. HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 8, 535–542 (2013).
Hung, R.-J., Spaeth, C.S., Yesilyurt, H.G. & Terman, J.R. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat. Cell Biol. 15, 1445–1454 (2013).
Lee, B.C. et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol. Cell 51, 397–404 (2013).
St. John, G. et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98, 9901–9906 (2001).
Tarrago, L., Kaya, A., Weerapana, E., Marino, S.M. & Gladyshev, V.N. Methionine sulfoxide reductases preferentially reduce unfolded oxidized proteins and protect cells from oxidative protein unfolding. J. Biol. Chem. 287, 24448–24459 (2012).
Ma, X.-X. et al. Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1. J. Biol. Chem. 286, 13430–13437 (2011).
Berg, J., Hung, Y.P. & Yellen, G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat. Methods 6, 161–166 (2009).
Luo, S. & Levine, R.L. Methionine in proteins defends against oxidative stress. FASEB J. 23, 464–472 (2009).
Tantama, M., Hung, Y.P. & Yellen, G. in Progress in Brain Research Vol. 196 (eds. Knöpfel, T. & Boyden, E.S.) 235–263 (Elsevier, 2012).
Schwarzländer, M. et al. The 'mitoflash' probe cpYFP does not respond to superoxide. Nature 514, E12–E14 (2014).
Hung, Y.P., Albeck, J.G., Tantama, M. & Yellen, G. Imaging cytosolic NADH-NAD+ redox state with a genetically encoded fluorescent biosensor. Cell Metab. 14, 545–554 (2011).
Mishina, N.M. et al. in Methods in Enzymology Vol. 526 (eds. Cadenas, E. & Packer, L.) 175–187 (Academic Press, 2013).
Poburko, D., Santo-Domingo, J. & Demaurex, N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 286, 11672–11684 (2011).
Ranaivoson, F.M. et al. Methionine sulfoxide reductase B displays a high level of flexibility. J. Mol. Biol. 394, 83–93 (2009).
Tarrago, L. et al. Plant thioredoxin CDSP32 regenerates 1-cys methionine sulfoxide reductase B activity through the direct reduction of sulfenic acid. J. Biol. Chem. 285, 14964–14972 (2010).
Vieira Dos Santos, C., Cuiné, S., Rouhier, N. & Rey, P. The Arabidopsis plastidic methionine sulfoxide reductase B proteins. Sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiol. 138, 909–922 (2005).
Patterson, G.H., Knobel, S.M., Sharif, W.D., Kain, S.R. & Piston, D.W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73, 2782–2790 (1997).
Tarrago, L. et al. Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxide reductases B by glutaredoxins and thioredoxins. J. Biol. Chem. 284, 18963–18971 (2009).
Chung, C.T., Niemela, S.L. & Miller, R.H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86, 2172–2175 (1989).
Acknowledgements
We thank P. Rey (Laboratoire d'Ecophysiologie Moléculaire des Plantes, UMR7265 CEA-CNRS-Aix-Marseille Université) for the kind gift of dabsyl-MetO and V. Belousov and V. Verkhusha for discussion. This study was supported by US National Institutes of Health grants AG021518 and GM065204 to V.N.G. and HL48743 to T.M.
Author information
Authors and Affiliations
Contributions
L.T. designed, created and characterized the sensors; performed experiments with E. coli cells; analyzed the data and wrote the paper. Z.P. performed experiments with HEK293 cells, analyzed the data and wrote the paper. B.C.L. performed experiments with MICAL1-oxidized actin and analyzed the data. T.M. contributed reagents and tools and analyzed the data. V.N.G. designed the sensors, supervised the research and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–15 and Supplementary Tables 1–4. (PDF 1887 kb)
Rights and permissions
About this article
Cite this article
Tarrago, L., Péterfi, Z., Lee, B. et al. Monitoring methionine sulfoxide with stereospecific mechanism-based fluorescent sensors. Nat Chem Biol 11, 332–338 (2015). https://doi.org/10.1038/nchembio.1787
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1787
This article is cited by
-
Metabolic changes induced by Cuscuta campestris Yunck in the host species Artemisia campestris subsp. variabilis (Ten.) Greuter as a strategy for successful parasitisation
Planta (2022)
-
Hypermethioninemia induces memory deficits and morphological changes in hippocampus of young rats: implications on pathogenesis
Amino Acids (2020)
-
A genetically encoded toolkit for tracking live-cell histidine dynamics in space and time
Scientific Reports (2017)
-
Intracellular repair of oxidation-damaged α-synuclein fails to target C-terminal modification sites
Nature Communications (2016)