Abstract
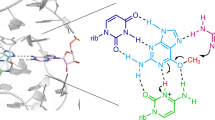
Uniquely among known ribozymes, the glmS ribozyme-riboswitch requires a small-molecule coenzyme, glucosamine-6-phosphate (GlcN6P). Although consistent with its gene-regulatory function, the use of GlcN6P is unexpected because all of the other characterized self-cleaving ribozymes use RNA functional groups or divalent cations for catalysis. To determine what active site features make this ribozyme reliant on GlcN6P and to evaluate whether it might have evolved from a coenzyme-independent ancestor, we isolated a GlcN6P-independent variant through in vitro selection. Three active site mutations suffice to generate a highly reactive RNA that adopts the wild-type fold but uses divalent cations for catalysis and is insensitive to GlcN6P. Biochemical and crystallographic comparisons of wild-type and mutant ribozymes show that a handful of functional groups fine-tune the RNA to be either coenzyme or cation dependent. These results indicate that a few mutations can confer new biochemical activities on structured RNAs. Thus, families of structurally related ribozymes with divergent function may exist.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A. & Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004).
Ferré-D'Amaré, A.R. The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA. Q. Rev. Biophys. 43, 423–447 (2010).
Collins, J.A., Irnov, I., Baker, S. & Winkler, W.C. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 21, 3356–3368 (2007).
Klein, D.J. & Ferré-D'Amaré, A.R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313, 1752–1756 (2006).
Hampel, K.J. & Tinsley, M.M. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry 45, 7861–7871 (2006).
Tinsley, R.A., Furchak, J.R. & Walter, N.G. trans-Acting glmS catalytic riboswitch: locked and loaded. RNA 13, 468–477 (2007).
Zhang, J., Lau, M.W. & Ferré-D'Amaré, A.R. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry 49, 9123–9131 (2010).
McCown, P.J., Roth, A. & Breaker, R. An expanded collection and refined consensus model of glmS ribozymes. RNA 17, 728–736 (2011).
Webb, C.-H.T., Riccitelli, N.J., Ruminski, D.J. & Lupták, A. Widespread occurrence of self-cleaving ribozymes. Science 326, 953 (2009).
de la Peña, M. & García-Robles, I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 16, 1943–1950 (2010).
Salehi-Ashtiani, K. & Szostak, J.W. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414, 82–84 (2001).
Williams, K.P., Ciafré, S. & Tocchini-Valentini, G.P. Selection of novel Mg2+-dependent self-cleaving ribozymes. EMBO J. 14, 4551–4557 (1995).
Tang, J. & Breaker, R.R. Structural diversity of self-cleaving ribozymes. Proc. Natl. Acad. Sci. USA 97, 5784–5789 (2000).
Lazarev, D., Puskarz, I.J. & Breaker, R.R. Substrate specificity and reaction kinetics of an X-motif ribozyme. RNA 9, 688–697 (2003).
Ferré-D'Amaré, A.R. & Scott, W.G. Small self-cleaving ribozymes. Cold Spring Harb. Perspect. Biol. 2, a003574 (2010).
Jayasena, V.K. & Gold, L. In vitro selection of self-cleaving RNAs with a low pH optimum. Proc. Natl. Acad. Sci. USA 94, 10612–10617 (1997).
Brooks, K.M. & Hampel, K.J. Rapid steps in the glmS ribozyme catalytic pathway: cation and ligand requirements. Biochemistry 50, 2424–2433 (2011).
Emilsson, G.M., Nakamura, S., Roth, A. & Breaker, R. Ribozyme speed limits. RNA 9, 907–918 (2003).
Rupert, P.B. & Ferré-D'Amaré, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410, 780–786 (2001).
Rupert, P.B., Massey, A.P., Sigurdsson, S.T. & Ferré-D'Amaré, A.R. Transition state stabilization by a catalytic RNA. Science 298, 1421–1424 (2002).
Bevilacqua, P.C. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry 42, 2259–2265 (2003).
Ferré-D'Amaré, A.R. The hairpin ribozyme. Biopolymers 73, 71–78 (2004).
Kath-Schorr, S. et al. General acid-base catalysis mediated by nucleobases in the hairpin ribozyme. J. Am. Chem. Soc. 134, 16717–16724 (2012).
Martick, M. & Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320 (2006).
Gong, B., Klein, D.J., Ferré-D'Amaré, A.R. & Carey, P.R. The glmS ribozyme tunes the catalytically critical pKa of its coenzyme glucosamine-6-phosphate. J. Am. Chem. Soc. 133, 14188–14191 (2011).
Davis, J.H., Dunican, B.F. & Strobel, S.A. glmS riboswitch binding to the glucosamine-6-phosphate α-anomer shifts the pKa toward neutrality. Biochemistry 50, 7236–7242 (2011).
Viladoms, J. & Fedor, M.J. The glmS ribozyme cofactor is a general acid-base catalyst. J. Am. Chem. Soc. 134, 19043–19049 (2012).
Viladoms, J., Scott, L. & Fedor, M. An active-site guanine participates in glmS ribozyme catalysis in its protonated state. J. Am. Chem. Soc. 133, 18388–18396 (2011).
Link, K.H., Guo, L. & Breaker, R. Examination of the structural and functional versatility of glmS ribozymes by using in vitro selection. Nucleic Acids Res. 34, 4968–4975 (2006).
Deigan, K.E. & Ferré-D'Amaré, A. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc. Chem. Res. 44, 1329–1338 (2011).
Posakony, J.J. & Ferré-D'Amaré, A.R. Glucosamine and glucosamine-6-phosphate derivatives: catalytic cofactor analogues for the glmS ribozyme. J. Org. Chem. 78, 4730–4743 (2013).
Ferré-D'Amaré, A.R. Use of a coenzyme by the glmS ribozyme-riboswitch suggests primordial expansion of RNA chemistry by small molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2942–2948 (2011).
Igloi, G.L. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27, 3842–3849 (1988).
Roth, A., Nahvi, A., Lee, M., Jona, I. & Breaker, R.R. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA 12, 607–619 (2006).
Klawuhn, K., Jansen, J.A., Souchek, J., Soukup, G.A. & Soukup, J.K. Analysis of metal ion dependence in glmS ribozyme self-cleavage and coenzyme binding. ChemBioChem 11, 2567–2571 (2010).
Cowan, J.A. Metallobiochemistry of RNA. Co(NH3)63+ as a probe for Mg2+ (aq) binding sites. J. Inorg. Biochem. 49, 171–175 (1993).
Draper, D.E. A guide to ions and RNA structure. RNA 10, 335–343 (2004).
Romani, A. & Scarpa, A. Regulation of cell magnesium. Arch. Biochem. Biophys. 298, 1–12 (1992).
Schatz, D., Leberman, R. & Eckstein, F. Interaction of Escherichia coli tRNASer with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc. Natl. Acad. Sci. USA 88, 6132–6136 (1991).
Jansen, J.A., McCarthy, T.J., Soukup, G.A. & Soukup, J.K. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat. Struct. Mol. Biol. 13, 517–523 (2006).
Cochrane, J.C., Lipchock, S.V. & Strobel, S.A. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 14, 97–105 (2007).
Klein, D.J., Wilkinson, S.R., Been, M.D. & Ferré-D'Amaré, A.R. Requirement of helix P2.2 and nucleotide G1 for positioning of the cleavage site and cofactor of the glmS ribozyme. J. Mol. Biol. 373, 178–189 (2007).
McCarthy, T.J. et al. Ligand requirements for glmS ribozyme self-cleavage. Chem. Biol. 12, 1221–1226 (2005).
Xiao, H., Murakami, H., Suga, H. & Ferré-D'Amaré, A.R. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature 454, 358–361 (2008).
Klein, D.J., Been, M.D. & Ferré-D'Amaré, A.R. Essential role of an active-site guanine in glmS ribozyme catalysis. J. Am. Chem. Soc. 129, 14858–14859 (2007).
Barrick, J.E. et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA 101, 6421–6426 (2004).
Soukup, G.A. Core requirements for glmS ribozyme self-cleavage reveal a putative pseudoknot structure. Nucleic Acids Res. 34, 968–975 (2006).
Schultes, E.A. & Bartel, D.P. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science 289, 448–452 (2000).
Nagano, N., Orengo, C.A. & Thornton, J.M. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321, 741–765 (2002).
Pitt, J.N. & Ferré-D'Amaré, A.R. Rapid construction of empirical RNA fitness landscapes. Science 330, 376–379 (2010).
Pitt, J.N. & Ferré-D'Amaré, A.R. Structure-guided engineering of the regioselectivity of RNA ligase ribozymes. J. Am. Chem. Soc. 131, 3532–3540 (2009).
Cochrane, J.C., Lipchock, S.V., Smith, K.D. & Strobel, S.A. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry 48, 3239–3246 (2009).
Baird, N.J. & Ferré-D'Amaré, A.R. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16, 598–609 (2010).
Klein, D.J. & Ferré-D'Amaré, A.R. Crystallization of the glmS ribozyme-riboswitch. Methods Mol. Biol. 540, 129–139 (2009).
Otwinowski, Z. & Minor, W. Processing of diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Brünger, A.T. et al. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
DeLano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, 2002).
Ryder, S.P., Ortoleva-Donnelly, L., Kosek, A.B. & Strobel, S.A. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 317, 92–109 (2000).
Acknowledgements
We thank the staff at beamlines 5.0.1 and 5.0.2 of the Advanced Light Source and of 12-ID (BESSRC CAT) of the Advanced Photon Source for crystallographic and SAXS data collection support, respectively; X. Fang and Y.-X. Wang for access to SAXS beamtime; D.-Y. Lee and R. Levine for access to MS; J. Sellers for access to a rapid quench apparatus; N. Baird for performing analysis of SAXS data; K. Deigan, J. Posakony and J. Zhang for discussions; and an anonymous referee for motivating the phosphorothioate interference analysis of glmSAAG and glmSUAG. M.W.L.L. was a recipient of the Croucher Foundation Fellowship. This work was supported in part by the intramural program of the US National Heart, Lung and Blood Institute–National Institutes of Health.
Author information
Authors and Affiliations
Contributions
M.W.L.L. and A.R.F.-D. designed experiments, M.W.L.L. performed all of the biochemistry, M.W.L.L. collected SAXS data and grew crystals and M.W.L.L. and A.R.F.-D. collected diffraction data, solved and refined the crystal structures and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–10 and Supplementary Tables 1–4. (PDF 12189 kb)
Rights and permissions
About this article
Cite this article
Lau, M., Ferré-D'Amaré, A. An in vitro evolved glmS ribozyme has the wild-type fold but loses coenzyme dependence. Nat Chem Biol 9, 805–810 (2013). https://doi.org/10.1038/nchembio.1360
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1360
This article is cited by
-
The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme
Nature Chemical Biology (2017)