Abstract
Many organs are composed of complex tissue walls that are structurally organized to optimize organ function. In particular, the ventricular myocardial wall of the heart comprises an outer compact layer that concentrically encircles the ridge-like inner trabecular layer. Although disruption in the morphogenesis of this myocardial wall can lead to various forms of congenital heart disease1 and non-compaction cardiomyopathies2, it remains unclear how embryonic cardiomyocytes assemble to form ventricular wall layers of appropriate spatial dimensions and myocardial mass. Here we use advanced genetic and imaging tools in zebrafish to reveal an interplay between myocardial Notch and Erbb2 signalling that directs the spatial allocation of myocardial cells to their proper morphological positions in the ventricular wall. Although previous studies have shown that endocardial Notch signalling non-cell-autonomously promotes myocardial trabeculation through Erbb2 and bone morphogenetic protein (BMP) signalling3, we discover that distinct ventricular cardiomyocyte clusters exhibit myocardial Notch activity that cell-autonomously inhibits Erbb2 signalling and prevents cardiomyocyte sprouting and trabeculation. Myocardial-specific Notch inactivation leads to ventricles of reduced size and increased wall thickness because of excessive trabeculae, whereas widespread myocardial Notch activity results in ventricles of increased size with a single-cell-thick wall but no trabeculae. Notably, this myocardial Notch signalling is activated non-cell-autonomously by neighbouring Erbb2-activated cardiomyocytes that sprout and form nascent trabeculae. Thus, these findings support an interactive cellular feedback process that guides the assembly of cardiomyocytes to morphologically create the ventricular myocardial wall and more broadly provide insight into the cellular dynamics of how diverse cell lineages organize to create form.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fahed, A. C., Gelb, B. D., Seidman, J. G. & Seidman, C. E. Genetics of congenital heart disease: the glass half empty. Circ. Res. 112, 707–720 (2013)
Zemrak, F. et al. The relationship of left ventricular trabeculation to ventricular function and structure over a 9.5-year follow-up: the MESA study. J. Am. Coll. Cardiol. 64, 1971–1980 (2014)
Grego-Bessa, J. et al. Notch signaling is essential for ventricular chamber development. Dev. Cell 12, 415–429 (2007)
Gupta, V. & Poss, K. D. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 484, 479–484 (2012)
Staudt, D. W. et al. High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation. Development 141, 585–593 (2014)
Ghabrial, A. S. & Krasnow, M. A. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature 441, 746–749 (2006)
Siekmann, A. F. & Lawson, N. D. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature 445, 781–784 (2007)
Clark, B. S. et al. Loss of Llgl1 in retinal neuroepithelia reveals links between apical domain size, Notch activity and neurogenesis. Development 139, 1599–1610 (2012)
Samsa, L. A. et al. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development 142, 4080–4091 (2015)
Liu, J. et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development 137, 3867–3875 (2010)
Ong, L. L., Kim, N., Mima, T., Cohen-Gould, L. & Mikawa, T. Trabecular myocytes of the embryonic heart require N-cadherin for migratory unit identity. Dev. Biol. 193, 1–9 (1998)
Parsons, M. J. et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 (2009)
Zhao, L. et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl Acad. Sci. USA 111, 1403–1408 (2014)
Anderson, R. M. et al. Hepatocyte growth factor signaling in intrapancreatic ductal cells drives pancreatic morphogenesis. PLoS Genet. 9, e1003650 (2013)
Ninov, N., Borius, M. & Stainier, D. Y. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 139, 1557–1567 (2012)
Mathews, E. S. et al. Mutation of 3-hydroxy-3-methylglutaryl CoA synthase I reveals requirements for isoprenoid and cholesterol synthesis in oligodendrocyte migration arrest, axon wrapping, and myelin gene expression. J. Neurosci. 34, 3402–3412 (2014)
Peshkovsky, C., Totong, R. & Yelon, D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev. Dyn. 240, 446–456 (2011)
Lyons, D. A. et al. erbb3 and erbb2 are essential for Schwann cell migration and myelination in zebrafish. Curr. Biol. 15, 513–524 (2005)
Collery, R. F. & Link, B. A. Dynamic smad-mediated BMP signaling revealed through transgenic zebrafish. Dev. Dyn. 240, 712–722 (2011)
Chen, H. et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131, 2219–2231 (2004)
Sternberg, P. W. Lateral inhibition during vulval induction in Caenorhabditis elegans. Nature 335, 551–554 (1988)
Heitzler, P. & Simpson, P. The choice of cell fate in the epidermis of Drosophila. Cell 64, 1083–1092 (1991)
Schumacher, J. A., Bloomekatz, J., Garavito-Aguilar, Z. V. & Yelon, D. tal1 regulates the formation of intercellular junctions and the maintenance of identity in the endocardium. Dev. Biol. 383, 214–226 (2013)
Rentschler, S. et al. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J. Clin. Invest. 121, 525–533 (2011)
Zhao, C. et al. Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation. Development 141, 281–295 (2014)
Yang, J. et al. Inhibition of Notch2 by Numb/Numblike controls myocardial compaction in the heart. Cardiovasc. Res. 96, 276–285 (2012)
Leimeister, C., Externbrink, A., Klamt, B. & Gessler, M. Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech. Dev. 85, 173–177 (1999)
Li, L. et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nature Genet. 16, 243–251 (1997)
Luxán, G. et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nature Med. 19, 193–201 (2013)
Chi, N. C. et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 22, 734–739 (2008)
Palencia-Desai, S. et al. Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development 138, 4721–4732 (2011)
D’Amico, L., Scott, I. C., Jungblut, B. & Stainier, D. Y. A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr. Biol. 17, 252–259 (2007)
Kikuchi, K. et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601–605 (2010)
Kettleborough, R. N. et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496, 494–497 (2013)
Huang, C. J., Tu, C. T., Hsiao, C. D., Hsieh, F. J. & Tsai, H. J. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30–40 (2003)
Thermes, V. et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 118, 91–98 (2002)
Mosimann, C. et al. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177 (2011)
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007)
Zhou, Y. et al. Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645–648 (2011)
Zhang, R. et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497–501 (2013)
Yu, P. B. et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nature Chem. Biol. 4, 33–41 (2008)
Acknowledgements
We thank N. Tedeschi for fish care; B. Le for experimental assistance; S. Evans, D. Yelon, and Chi laboratory members for comments on the manuscript; N. Ninov and D. Stainier for plasmids; B. Link for the d2GFP BMP and d2GFP Notch reporter lines; N. Lawson for the eGFP Notch reporter line; B. Appel for the myocardial Cerulean line; K. Poss for the myocardial CreER and Brainbow/priZm lines; and W. Talbot for the erbb2 mutant. This work was supported in part by grants from American Heart Association (14POST20380738) to L.Z.; the March of Dimes (1-FY14-327) to R.A.M.; the NIH/NHLBI (5R01HL127067) to C.G.B. and C.E.B.; and the National Institutes of Health to N.C.C.
Author information
Authors and Affiliations
Contributions
P.H. and N.C.C. conceived the project and the design of the experimental strategy. P.H., J.R., J.B., R.Z., and J.D.G. conducted experiments. L.Z. generated the ubi:RSdnM transgenic line. P.H. and N.C.C. generated and characterized the myl7:Cre transgenic line. C.E.B., C.G.B., and R.A.M. provided key reagents. P.H., J.B. and N.C.C. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks B. G. Bruneau and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Notch signalling is dynamically activated in the endocardium and myocardium during heart development.
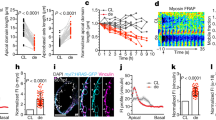
a–f, Confocal slices of Tg(Tp1:d2GFP; myl7:mCherry) hearts reveal that Notch signalling is in the ventricular endocardium (yellow arrows) but not in the myocardium at 24 hpf (n = 11) and 36 hpf (n = 8), but (g–i) becomes restricted to the AV and OFT endocardium by 48 hpf (n = 12). j–o, Tg(Tp1:d2GFP; kdrl:ras–mCherry) confocal imaging confirms that Tp1:d2GFP is expressed in the ventricular endocardium at (j–l) 24 hpf (n = 8) but becomes localized to the AV or OFT endocardium as well as non-endocardial cells in the outer ventricular myocardial wall (white arrows) by (m–o) 96 hpf (n = 10). p, q, Three-dimensional confocal reconstructions of the (p) exterior and (q) interior regions of 72 hpf Tg(Tp1:d2GFP; myl7:mCherry) hearts reveal that Notch-activated Tp1:d2GFP+ cells are present in cardiomyocyte clusters (green, numbers in parentheses) and excluded from nascent cardiac trabeculae (pseudo-colour magenta, numbers). r, Graph shows that the number of cardiac trabeculae (x axis) and Tp1:d2GFP+ cardiomyocyte clusters (y axis) are similar within the ventricle (n = 30) at 72 hpf. Size of dots indicates the number of embryos with a particular number of trabeculae and Tp1:d2GFP+ clusters. Line represents a linear regression fitted to the data. s, t, Myocardial anti-MHC/MF20 immunostaining of Tg(Tp1:d2GFP) hearts reveals a loss of myocardial Tp1:d2GFP Notch reporter signal at 30 and 90 dpf hearts (n = 5 hearts per stage). White arrows, likely Tp1:d2GFP+ cardiomyocytes; yellow arrows, Tp1:d2GFP+ endocardial cells; white and yellow asterisks, AV and OFT. Dashed line in s outlines ventricle. V, ventricle; A, atrium. Scale bar, 25 μm.
Extended Data Figure 2 DAPT treatment validates that the Notch reporter Tp1:d2GFP monitors dynamic Notch signalling more closely than Tp1:eGFP, and reveals opposing roles of Notch signalling on trabeculation at different developmental stages.
a–d, At 48hpf, (a) Tp1:d2GFP expression is restricted to the AV and OFT endocardium (n = 8/8 embryos) whereas (c) Tp1:eGFP is expressed in the ventricular, AV and OFT endocardium (n = 6/6). However, gfp mRNA is primarily expressed in the AV and OFT regions in both (b) Tg(Tp1:d2GFP; myl7:mCherry) (n = 10/10) and (d) Tg(Tp1:eGFP; myl7:mCherry) embryos (n = 5/5), revealing that Tp1:d2GFP expression most closely matches Notch reporter activity. e–h, After 24 h DAPT treatments of (e, f) Tg(Tp1:d2GFP; myl7:H2A–mCherry) and (g, h) Tg(Tp1:eGFP; myl7:H2A–mCherry) embryos at 72 hpf, (f) Tp1:d2GFP is more diminished throughout the heart at 96 hpf (n = 8/10) compared with (h) Tp1:eGFP (n = 6/7), confirming Tp1:d2GFP signal more faithfully recapitulates Notch signalling dynamics. m–p, Tg(Tp1:d2GFP; myl7:mCherry) hearts DAPT-treated from 60 to 72 hpf exhibit increased trabeculation (white arrowheads) and diminished Tp1:d2GFP Notch reporter activity (n = 12/16) than (i–l) DMSO-treated hearts (n = 0/20). However, (r) Tg(myl7:mCherry) hearts DAPT-treated from 20 to 48 hpf exhibit reduced trabeculae at 120 hpf (n = 12/15) than (q) DMSO-treated hearts (n = 0/20). s, Graph represents trabeculae/total ventricular area in embryos treated with DMSO or DAPT in q and r. White and yellow arrows, myocardial and endocardial Notch reporter activity; white arrowheads, trabeculae; white and yellow asterisks, AV and OFT. Scale bar, 25 μm. Mean ± s.e.m. *P < 0.05 by Student’s t-test.
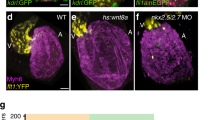
Extended Data Figure 3 Tp1:eGFP labels the ventricular outer wall during early cardiac development, which becomes the distinctive ventricular primordial myocardium in adults.
Using the Tp1:eGFP Notch reporter, which exhibits greater fluorescence perdurance than Tp1:d2GFP, we performed limited fate mapping of Notch activated cardiac cells during ventricular morphogenesis. a, b, Tp1:eGFP is expressed not only in ventricular cardiomyocytes (red nuclei, white arrows) at 72 hpf but also throughout the ventricular endocardium because of eGFP perdurance (yellow arrows) (n = 12). c, d, Although diminishing in the ventricular endocardium (yellow arrows) at 96 hpf (n = 14), Tp1:eGFP expands in the outer ventricular myocardial wall (white arrows), yet is notably absent from myocardial trabeculae (white arrowheads). e, f, By 30 and 45 dpf (n = 6, n = 5), Tp1:eGFP remains in the peripheral ventricular (primordial) myocardial layer, which is one cardiomyocyte thick (myl7:H2A–mCherry+/red and MF20+/blue), but is reduced in the ventricular but not the AV or OFT endocardium. g–i, At 60 dpf (n = 5), (h) new cardiomyocytes (cortical layer, yellow arrowheads) form over the Tp1:eGFP+ primordial myocardium (white arrows) at the ventricular myocardial base (yellow box in g) and extend towards the apex where (i)Tp1:eGFP+ cardiomyocytes (white arrows) still remain the outer most layer of the ventricular myocardium (white box in g). j, However, by 90 dpf (n = 5), this new cortical myocardial layer (yellow arrowheads) spreads over the apical Tp1:eGFP+ ventricular primordial myocardium (white arrows). k–m, In adult hearts (90 dpf), Tp1:eGFP is primarily found in the (k, n = 5) myl7:H2A–mCherry+ primordial myocardium but not in the (l, n = 5) endocardium marked by kdrl:ras–mCherry, nor (m, n = 3) epicardium marked by Raldh2 localization. n–t, Adult hearts (6 months) were further examined to assess the cellular attributes of the primordial layer. n, Anti-MHC/MF20 immunostaining confirms that Tp1:eGFP+ cardiac cells are myocardial (n = 5). o, Anti-α-actinin immunostaining reveals that trabecular (white arrowheads) and cortical (yellow arrowheads) cardiomyocytes display organized sarcomeric structures but the Tp1:eGFP+ primordial cardiomyocytes (arrows) do not (n = 7). p–t, Wheat germ agglutinin (WGA) staining shows that (p, q) the Tp1:eGFP+ primordial myocardial layer is surrounded by extensive extracellular matrix (n = 5) and that (r–t) Tg(myl7:ras–eGFP) primordial cardiomyocytes display a thin cellular morphology compared with other ventricular cardiomyocytes (n = 10). q, An X–Z reconstruction of confocal stacks from Tp1:eGFP and wheat germ agglutinin stainings at the dashed line shown in p. b, d, h–i, t, Magnifications of the boxed areas in a, c, g, s, respectively. White and yellow arrows, myocardial and endocardial Tp1:eGFP; white and yellow arrowheads, trabeculae and cortical layer; white and yellow asterisks, AV and OFT. Scale bar, 25 μm.
Extended Data Figure 4 BMP signalling, which marks trabeculae, is required for expanding but not initiating trabeculae formation and has no effect on myocardial Notch activity.
a–l, Tg(BRE:d2GFP; myl7:mCherry) hearts were treated with (a–c) DMSO, (d–f) DAPT, (g–i) AG1478, or (j–l) Dorsomorphin at 60 hpf and imaged at 72 hpf. a–c, DMSO-treated hearts express the BRE:d2GFP BMP reporter in trabeculae (arrowheads) and in the AV myocardium (yellow arrows, n = 11/11 embryos). d–f, DAPT-treated hearts exhibit increased trabeculation and BRE:d2GFP expression in these forming trabeculae (arrowheads, n = 9/12). g–i, AG1478-treated hearts fail to form trabeculae (n = 9/10) and only express the BRE:d2GFP BMP reporter in the AV myocardium (yellow arrow). j–l, Dorsomorphin-treated hearts form cardiac trabeculae (arrowheads) but fail to express the BRE:d2GFP BMP reporter in both cardiac trabeculae and the AV myocardium (n = 10/12). m–p, Treating Tg(Tp1:d2GFP; myl7:mCherry) embryos with Dorsomorphin from 60 to 72 hpf did not affect the initiation of trabeculae (arrowheads) nor the activation of myocardial Notch signalling (white arrows, n = 13/16) compared with treating with DMSO (see Extended Data Fig. 2i–l). q, r, Although Tg(myl7:mCherry) hearts treated with (q) DMSO or (r) Dorsomorphin from 60 hpf to 7 dpf form similar numbers of trabeculae (arrowheads), Dorsomorphin-treated hearts display trabeculae that are stunted/reduced in size (n = 12/15) compared with DMSO-treated control hearts (n = 0/15). s, Graph reveals a significant reduction in the trabecular/ventricular area ratio in Dorsmorphin-treated fish compared with DMSO-treated controls. Arrowheads, trabeculae; yellow arrows, AV myocardium; white arrows, Tp1:d2GFP+ myocardium. White asterisks, AV. Mean ± s.e.m. *P < 0.05 by Student’s t-test. Scale bar, 25 μm.
Extended Data Figure 5 Altering myocardial Notch signalling affects ventricular size and wall thickness but not total number of ventricular cardiomyocytes.
a–d, The Tg(myl7:Cre) transgenic line used to specifically perturb Notch signalling in the myocardium was validated by confirming that Cre expression is restricted to the myocardium. Activity of myl7:Cre, as visualized by (c) GFP expression from the switch line, β-act2:RSG, exclusively overlaps with (b, d) myl7:Cerulean expression at 120 hpf (n = 10 embryos). Quantitative analyses of (e) ventricular size and (f) wall thickness performed on confocal images from Fig. 2a–h reveal that myocardial Notch signalling restricts ventricular size while promoting ventricular wall thickness. e, Ventricular size measurements were normalized to respective controls for each condition. f, Individual measurements (dots) of myocardial thickness were taken across the outer curvature of the ventricle (n = 30 measurements, 6 measurements were taken per embryo, 5 embryos per condition). Dashed line represents the ventricular wall thickness that distinguishes trabeculated myocardial thickness from ventricular outer wall myocardial thickness in control hearts. Crosses denote mean and s.e.m. g–p, Quantitative analysis of (g) trabecular cardiomyocytes and (p) total ventricular cardiomyocytes was calculated by counting myocardial nuclei labelled with myl7:H2A–mCherry or anti-Mef2 immunostaining using embryos from Fig. 2i–p for g, or from three-dimensional reconstructions in h–o for p. In g, the number of trabecular/total ventricular cardiomyocytes was used to calculate the percentage of trabecular cardiomyocytes for each condition. In p, total ventricular cardiomyocytes were normalized to respective controls for each condition. n, Number of embryos analysed per condition. Mean ± s.e.m. *P < 0.05 by Student’s t-test. NS, not significant. Scale bar, 25 μm.
Extended Data Figure 6 Myocardial Notch activation can inhibit the formation and expansion of cardiac trabeculae at various cardiac developmental stages.
a–g, Tg(myl7:Cre; hsp70l:RSN) and Tg(hsp70l:RSN) (control) embryos were heat-shocked (HS) during various developmental time windows as indicated and imaged at 7 dpf to assess the effects of constitutive myocardial Notch signalling on cardiac trabeculae formation. a, Red arrows in schematic indicate the time points at which embryos in the corresponding panels were heat-shocked. b, Control Tg(hsp70l:RSN) embryos heat-shocked from 60 hpf to 7 dpf ubiquitously express mCherry but do not overexpress myocardial NICD. They form cardiac trabeculae (arrowheads) similar to wild-type embryos (control, n = 14/15). c, However, Tg(myl7:Cre; hsp70l:RSN) embryos heat-shocked from 60 hpf to 7 dpf overexpress NICD-P2A–Emerald throughout the myocardium and fail to form cardiac trabeculae (n = 9/12). Although Tg(myl7:Cre; hsp70l:RSN) embryos heat-shocked at (d) 80 hpf, (e) 96 hpf, and (f) 120 hpf form trabeculae, these embryos exhibit stunted/smaller trabeculae after heat-shocking (n = 9/10, 10/14, and 12/16, respectively). g, Graph of trabeculae/total ventricular area of heat-shocked embryos from b–f, showing that myocardial Notch over-activation inhibits the progression of cardiac trabeculae formation. h, i, Although heat-shocking Tg(myl7:Cre; hsp70l:RSN) from 60 to 120 hpf initially inhibits trabeculae formation, (i) the ventricular myocardium (detected by anti-MHC/MF20 immunostaining, magenta) can still form trabeculae, albeit at reduced numbers (n = 4/5) by 30 dpf after stopping NICD overexpression compared with (h) heat-shocked Tg(hsp70l:RSN) hearts (control, n = 0/8). HS, heat-shock; white arrowheads, trabeculae. Scale bar, 25 μm. Mean ± s.e.m. *P < 0.05 by Student’s t-test.
Extended Data Figure 7 Notch signalling regulates cardiomyocyte cell junctions during cardiac trabeculae formation.
a–d, In DMSO-treated (control) 72 hpf wild-type hearts, N-cadherin is localized at cell junctions of cardiomyocytes within the ventricular outer wall (arrows) but redistributes away from these cell–cell contacts in cardiomyocytes that extend into the lumen to form trabeculae (arrowheads) (n = 12/12). e–h, Notch inhibition by DAPT treatment promotes N-cadherin redistribution and results in increased trabeculation (n = 8/11). m–p, Conversely, myocardial Notch activation by heat shocking (HS) Tg(myl7:Cre; hsp70l:RSN) leads to diminished N-cadherin redistribution and reduced trabeculation (n = 7/10) compared with (i–l) heat-shocked Tg(hsp70l:RSN) control hearts (n = 0/10). Nascent cardiac trabeculae were pseudo-coloured magenta in c, g and k. b, d, f, h, j, l, n, p, Magnifications of boxed areas in a, c, e, g, i, k, m, o, respectively. Arrowheads, N-cadherin redistributed from cell–cell contacts; arrows, N-cadherin at cell–cell contacts within outer wall. Scale bar, 25 μm.
Extended Data Figure 8 Tamoxifen treatment of Tg(myl7:CreER; priZm) embryos at 48 hpf labels adjacent individual cardiomyocytes with combinations of distinct fluorescent colours.
Tg(myl7:CreER; priZm) embryos were treated with 4-HT at 48 hpf and confocal imaged at 60 hpf before the initiation of cardiomyocytes forming trabeculae. Individual cardiomyocytes (arrowheads) are labelled with distinct combinations of fluorescent proteins allowing for tracking of specific cardiomyocyte clones (n = 6). White arrowheads, cardiomyocytes; V, ventricle; A, atrium; white asterisk, AV. Scale bar, 25 μm.
Extended Data Figure 9 Notch and Erbb2 signalling pathways form a feedback loop during cardiac trabeculation.
a, b, Compared with (a) DMSO-treated Tg(Tp1:d2GFP; myl7:mCherry) (controls) embryos, (b) inhibiting Erbb2 function with AG1478 from 60 to 72 hpf blocks trabeculation and myocardial Notch signalling (n = 14/17), confirming erbb2 MO and mutant phenotypes. c, However, Notch inhibition using DAPT cannot reverse the AG1478/Erbb2 inhibition effect on trabeculae formation (n = 11/12). d, e, Consistent with these results, (d) control MO-injected Tg(hsp70l:dnM; myl7:mCherry) embryos expressing heat-shock induced dnMAML from 60 to 72 hpf display increased trabeculation (arrowheads, n = 9/11); (e) however, erbb2 MO-injected embryos expressing heat-shock induced dnMAML fail to display trabeculae (n = 9/12) as similarly observed in erbb2 MO-injected embryos alone (Fig. 3). f–j, The erbb2 fluorescent in situ hybridization and GFP co-immunostaining performed on 72 hpf Tg(Tp1:d2GFP) hearts reveal that erbb2 is expressed in an intermittent pattern across the ventricular wall and is specifically diminished in Tp1:d2GFP+ cells (arrows) (n = 6/6). l, p, Heat-shocked (HS) Tg(myl7:Cre; hsp70l:RSN) hearts, which exhibit constitutively activated myocardial Notch signalling (NICD) from 60 to 120 hpf, minimally express erbb2 in the myocardium (n = 8/11) compared with (k, o) heat-shocked Tg(hsp70l:RSN) control hearts (n = 0/20) at 120 hpf. Compared with (m, q) DMSO-treated control hearts (n = 0/10), (n, r) Notch-inhibited hearts by DAPT treatment from 60 to 72 hpf exhibit increased myocardial erbb2 expression as well as more trabeculae at 72 hpf (n = 8/10), supporting the idea that Notch signalling inhibits erbb2 expression. h, i–j, Magnifications of boxed areas in g, h, respectively. Arrowheads, trabeculae; arrows, Tp1:d2GFP+ cardiomyocytes; white and yellow asterisks, AV and OFT. Scale bar, 25 μm.
Extended Data Figure 10 Transplanted wild-type cardiomyocytes non-cell-autonomously activate Notch signalling in erbb2 morphant host cardiomyocytes.
a, On the basis of mosaic embryo studies from Fig. 3f–i, wild-type donor cardiomyocytes contribute equally to the outer ventricular wall (14/26 clones) or the trabeculae (12/26 clones) when transplanted into control MO host embryos (n = 12 embryos). However, when wild-type donor cells are transplanted into erbb2 MO host embryos (n = 10 embryos), they contribute more to the trabecular layer (19/23 clones) than to the ventricular outer wall (4/23 clones, P < 0.05 by Fisher’s exact test). b, On the basis of mosaic embryo studies from Fig. 3f–i, transplanting wild-type donor cells increases the number of erbb2 MO host cardiomyocytes expressing Tp1:d2GFP (n = 10 embryos) compared with non-transplanted erbb2 MO embryos (n = 16 embryos), but had no effect on the number of control MO host cells expressing Tp1:d2GFP (n = 12 embryos) compared with non-transplanted controls (n = 11 embryos). c, Quantitative data for Fig. 3f–i reveal that transplanted wild-type donor cardiomyocytes are primarily adjacent to host Tp1:d2GFP+ cardiomyocytes in erbb2 MO hearts (n = 10 embryos). Mean ± s.e.m. *P < 0.05 by Student’s t-test. NS, not significant.
Rights and permissions
About this article
Cite this article
Han, P., Bloomekatz, J., Ren, J. et al. Coordinating cardiomyocyte interactions to direct ventricular chamber morphogenesis. Nature 534, 700–704 (2016). https://doi.org/10.1038/nature18310
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18310
This article is cited by
-
Endothelial deletion of PTBP1 disrupts ventricular chamber development
Nature Communications (2023)
-
Nono deficiency impedes the proliferation and adhesion of H9c2 cardiomyocytes through Pi3k/Akt signaling pathway
Scientific Reports (2023)
-
Modeling of large-scale hoxbb cluster deletions in zebrafish uncovers a role for segmentation pathways in atrioventricular boundary specification
Cellular and Molecular Life Sciences (2023)
-
Functional screening of congenital heart disease risk loci identifies 5 genes essential for heart development in zebrafish
Cellular and Molecular Life Sciences (2023)
-
Cardiac cell type-specific responses to injury and contributions to heart regeneration
Cell Regeneration (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.