Abstract
Homozygosity has long been associated with rare, often devastating, Mendelian disorders1, and Darwin was one of the first to recognize that inbreeding reduces evolutionary fitness2. However, the effect of the more distant parental relatedness that is common in modern human populations is less well understood. Genomic data now allow us to investigate the effects of homozygosity on traits of public health importance by observing contiguous homozygous segments (runs of homozygosity), which are inferred to be homozygous along their complete length. Given the low levels of genome-wide homozygosity prevalent in most human populations, information is required on very large numbers of people to provide sufficient power3,4. Here we use runs of homozygosity to study 16 health-related quantitative traits in 354,224 individuals from 102 cohorts, and find statistically significant associations between summed runs of homozygosity and four complex traits: height, forced expiratory lung volume in one second, general cognitive ability and educational attainment (P < 1 × 10−300, 2.1 × 10−6, 2.5 × 10−10 and 1.8 × 10−10, respectively). In each case, increased homozygosity was associated with decreased trait value, equivalent to the offspring of first cousins being 1.2 cm shorter and having 10 months’ less education. Similar effect sizes were found across four continental groups and populations with different degrees of genome-wide homozygosity, providing evidence that homozygosity, rather than confounding, directly contributes to phenotypic variance. Contrary to earlier reports in substantially smaller samples5,6, no evidence was seen of an influence of genome-wide homozygosity on blood pressure and low density lipoprotein cholesterol, or ten other cardio-metabolic traits. Since directional dominance is predicted for traits under directional evolutionary selection7, this study provides evidence that increased stature and cognitive function have been positively selected in human evolution, whereas many important risk factors for late-onset complex diseases may not have been.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

 , on 16 traits.
, on 16 traits.
Similar content being viewed by others
References
Garrod, A. The incidence of alkaptonuria: a study of chemical individuality. Lancet 160, 1616–1620 (1902)
Darwin, C. The Variation of Animals and Plants Under Domestication (Appleton, 1868)
McQuillan, R. et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83, 359–372 (2008)
McQuillan, R. et al. Evidence of inbreeding depression on human height. PLoS Genet. 8, e1002655 (2012)
Rudan, I. et al. Inbreeding and the genetic complexity of human hypertension. Genetics 163, 1011–1021 (2003)
Campbell, H. et al. Effects of genome-wide heterozygosity on a range of biomedically relevant human quantitative traits. Hum. Mol. Genet. 16, 233–241 (2007)
Charlesworth, D. & Willis, J. H. The genetics of inbreeding depression. Nature Rev. Genet. 10, 783–796 (2009)
Wright, S. Evolution and the Genetics of Populations, Vol. 3: Experimental Results and Evolutionary Deductions (University of Chicago Press, 1977)
Wright, S. Coefficients of inbreeding and relationships. Am. Nat. 56, 330–339 (1922)
Broman, K. W. & Weber, J. L. Long homozygous chromosomal segments in reference families from the Centre d’Étude du Polymorphisme Humain. Am. J. Hum. Genet. 65, 1493–1500 (1999)
Gibson, J., Morton, N. E. & Collins, A. Extended tracts of homozygosity in outbred human populations. Hum. Mol. Genet. 15, 789–795 (2006)
Kirin, M. et al. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE 5, e13996 (2010)
Keller, M. C., Visscher, P. M. & Goddard, M. E. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics 189, 237–249 (2011)
Pemberton, T. J. & Rosenberg, N. A. Population-genetic influences on genomic estimates of the inbreeding coefficient: a global perspective. Hum. Hered. 77, 37–48 (2014)
Abdellaoui, A. et al. Educational attainment influences levels of homozygosity through migration and assortative mating. PLoS ONE 10, e0118935 (2015)
Neel, J. V. et al. The effects of parental consanguinity and inbreeding in Hirado, Japan. II. Physical development, tapping rate, blood pressure, intelligence quotient, and school performance. Am. J. Hum. Genet. 22, 263–286 (1970)
Marioni, R. E. et al. Common genetic variants explain the majority of the correlation between height and intelligence: the generation Scotland study. Behav. Genet. 44, 91–96 (2014)
Wood, A. R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genet. 46, 1173–1186 (2014)
Deary, I. J. et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature 482, 212–215 (2012)
Morton, N. E. Effect of inbreeding on IQ and mental retardation. Proc. Natl Acad. Sci. USA 75, 3906–3908 (1978)
Najmabadi, H. et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63 (2011)
Nalls, M. A. et al. Measures of autozygosity in decline: globalization, urbanization, and its implications for medical genetics. PLoS Genet. 5, e1000415 (2009)
Flynn, J. R. Massive IQ gains in 14 nations: what IQ tests really measure. Psychol. Bull. 101, 171–191 (1987)
Galton, F. Natural inheritance (MacMillan, 1889)
Hoffman, J. I. et al. High-throughput sequencing reveals inbreeding depression in a natural population. Proc. Natl Acad. Sci. USA 111, 3775–3780 (2014)
Wright, A., Charlesworth, B., Rudan, I., Carothers, A. & Campbell, H. A polygenic basis for late-onset disease. Trends Genet. 19, 97–106 (2003)
Weiss, L. A., Pan, L., Abney, M. & Ober, C. The sex-specific genetic architecture of quantitative traits in humans. Nature Genet. 38, 218–222 (2006)
Powell, J. E. et al. Congruence of additive and non-additive effects on gene expression estimated from pedigree and SNP data. PLoS Genet. 9, e1003502 (2013)
Keller, M. C. et al. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 8, e1002656 (2012)
Darwin, C. The Effects of Crossing and Self Fertilization in the Vegetable Kingdom (John Murray, 1876)
Purcell, S. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Duijn, C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007)
Ronnegard, L., Shen, X. & Alam, M. hglm: a package for fitting hierarchical generalized linear models. R Journal 2, 20–28 (2010)
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010)
Ehret, G. B. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011)
Scott, R. A. et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature Genet. 44, 991–1005 (2012)
Willer, C. J. et al. Discovery and refinement of loci associated with lipid levels. Nature Genet. 45, 1274–1283 (2013)
Soler Artigas, M. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nature Genet. 43, 1082–1090 (2011)
Rietveld, C. A. et al. GWAS of 126,559 individuals identified genetic variants associated with educational attainment. Science 340, 1467–1471 (2013)
Stefansson, H. et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505, 361–366 (2014)
An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Aulchenko, Y. S., de Koning, D. J. & Haley, C. Genomewide rapid association using mixed model and regression: a fast and simple method for genome-wide pedigree-based quantitative trait loci association analysis. Genetics 177, 577–585 (2007)
Marioni, R. E. et al. Molecular genetic contributions to socioeconomic status and intelligence. Intelligence 44, 26–32 (2014)
Hedges, L. V. & Olkin, I. Statistical Methods for Meta-Analysis (Academic Press, New York, 1985)
Acknowledgements
This paper is the work of the ROHgen consortium. We thank the participants in all ROHgen studies; cohort-specific acknowledgements are detailed in Supplementary Table 6. This work was funded by a UK Medical Research Council (MRC) PhD studentship to P.K.J.; and J.F.W. and O.P. acknowledge support from the MRC Human Genetics Unit “QTL in Health and Disease” programme. We thank W. G. Hill for discussions and comments on the manuscript and K. Lindsay for administrative assistance.
Author information
Authors and Affiliations
Consortia
Contributions
C.Hal., P.N., M.Me., H.B., N.J.S., D.C., D.A.M., R.S.C., P.F., G.P., S.F.G., H.H., L.F., R.A.S., A.D.M., C.N.P., G.De., P.D., L.B., U.L., S.I.B., C.M.L., N.J.T., A.Ton., P.B.M., T.I.S., C.N.R., D.K.A., A.J.O., S.L.K., B.B., G.Ga., A.P.M., J.G.E., M.J.W., N.G.M., S.C.H., J.M.S., I.J.D., L.R.G., H.T., N.Pi., J.Ka., N.J.W., L.P., J.G.W., G.Gi., M.J.C., O.R., D.D.B., C.Gi., P.v.d.H., A.A.H., P.Kr., J.S., P.Kn., M.J., P.K.M., A.H., R.Sc., I.B.B., E.Va., D.M.B., D.B., K.L.M., M.B., C.M.v.D., D.K.S., A.Te., E.Z., A.Me., P.G., S.U., C.O., D.T., G.D.S., I.R., D.J.P., M.C., T.D.S., C.Hay., J.D., R.J.L., A.F.W., G.R.C., P.V., A.Sh., P.M.R., J.I.R., N.S., U.G., K.E.N., M.P., B.M.P., D.R.W., M.La., V.G., A.Ta., J.C.C., J.S.K., D.P.S., H.C., J.N.H., M.P., O.P. and J.F.W. designed individual studies. T.N., J.D.F., S.E., V.V., S.Tr., D.I.C., S.S.N., M.Ma., D.R., A.F., L.R.Y., E.H., C.Bo., J.R.P., S.C., U.B., G.M., T.Li., I.D., J.Z., J.P.B., E.S., S.Y., M.A.A., S.J.B., G.R.B., E.P.B., A.Ca., Y. Chan, S.J.C., Y.D.I.C., F.S.C., J.C., A.Co., L.Cu., G.Da., M.D., S.B.E., B.F., M.F.F., I.F., C.S.F., T.M.F., N.Fri., F.Ge., I.Gi., O.G., F.Gr., C.Gu., C.J.H., S.E.H., N.D.H., N.L.H., K.H., L.J.H., G.Ho., P.G.H., E.I., Å.J., P.J., J.J., M.Ka., S.K., S.M.K., N.M.K., H.K.K., M.Ku., J.Ku., J.L., R.A.L., T.Le., D.C.L., L.Li., M.L.L., A.Lo., T.Lu., A.Lu., S.M., K.M., J.B.M., C.Mei., T.M., C.Men., F.D.M., L.M., G.W.M., R.H.M., R.N., M.N., M.S.N., G.T.O., A.O., S.P., W.R.P., J.S.P., I.Pa., K.P., N.Po., S.Ra., P.R., S.S.R., H.R., A.R., L.M.R., R.R., B.Sa., R.M.S., V.S., A.Sa., L.J.S., S.Se., P.S., B.H.S., N.Sor., A.V.St., M.G.S., K.S., N.Ta., K.D.T., B.O.T., A.Tog., M.To., J.T., A.G.U., A.v.H.V., T.V., S.V., E.Vl., E.Vu., M.W., J.B.W., S.W., G.W., C.S.Y., G.Z., X.Z., M.Me., H.B., N.J.S., D.C., D.A.M., R.S.C., G.P., S.F.G., H.H., L.F., R.A.S., G.De., P.D., L.B., U.L., S.I.B., G.D.S., N.J.T., A.Ton., P.B.M., T.I.S., C.N.R., D.K.A., A.J.O., S.L.K., B.B., M.K.K., G.Ga., J.G.E., M.J.W., N.G.M., S.C.H., J.M.S., I.J.D., L.R.G., J.Ka., N.J.W., L.P., J.G.W., G.Gi., M.J.C., O.R., D.D.B., C.Gi., P.v.d.H., A.A.H., P.Kr., J.S., P.Kn., M.J., P.K.M., A.H., R.Sc., I.B.B., E.Va., D.M.B., D.B., K.L.M., M.B., C.M.v.D., D.K.S., E.Z., A.Me., P.G., C.O., D.T., D.J.P., M.C., T.D.S., C.Hay., R.J.L., A.F.W., G.R.C., P.V., A.Sh., P.M.R., J.I.R., N.S., U.G., M.P., B.M.P., D.R.W., M.La., J.C.C., J.S.K., D.P.S., J.N.H., M.P., O.P. and J.F.W. collected the data. S.Tr., D.I.C., M.C.C., C.Bo., U.B., I.D., M.A., F.W.A., S.J.B., D.J.B., E.B., E.P.B., A.Cc., S.J.C., J.C., I.F., T.M.F., C.Gu., C.J.H., T.B.H., N.D.H., M.I., E.I., J.J., P.Ko., M.Ku., L.J.L., R.A.L., L.Li., R.A.M., K.M., J.B.M., G.W.M., R.H.M., P.A.P., K.P., S.S.R., R.R., H.S., P.S., B.H.S., N.Sor., N.Sot., D.Va., J.B.W., C.S.Y., M.Me., N.J.S., D.C., D.A.M., R.S.C., P.F., G.P., S.F.G., H.H., L.F., G.De., P.D., L.B., U.L., S.I.B., C.M.L., A.Ton., P.B.M.,.C.N.R., D.K.A., A.J.O., S.L.K., B.B., G.Ga., A.P.M., M.J.W., N.G.M., S.C.H., J.M.S., I.J.D., L.R.G., J.Ka., N.J.W., L.P., M.J.C., D.D.B., P.v.d.H., P.Kr., M.J., P.K.M., A.H., R.Sc., I.B.B., D.M.B., D.B., K.L.M., M.B., C.M.v.D., D.K.S., E.Z., A.Me., P.G., S.U., C.O., I.R., D.J.P., M.C., T.D.S., C.Hay., A.F.W., G.R.C., P.V., A.Sh., P.M.R., J.I.R., N.S., U.G., K.E.N., B.M.P., D.R.W., M.La., V.G., D.P.S., H.C., O.P. and J.F.W. contributed to funding. P.K.J., T.E., H.Ma., N.E., I.Ga., T.N., A.U.J., C.Sc., A.V.Sm., W.Zhan., Y.O., A.Stc., J.D.F., W. Zhao, T.M.B., M.P.C., N.Fra., S.E., V.V., S.Tr., X.G., D.I.C., J.R.O., T.C., S.S.N., Y. Chen, M.Ma., D.R., M.Ta., A.F., T.Kac., A.Bj., A.v.d.S., Y.W., A.K.G., L.R.Y., L.W., E.H., C.A.R., O.M., M.C.C., C.P., N.V., C.Ba., A.A.A., H.R.W., D.Vu., H.Me., J.R.P., S.S.Mi., M.C.B., S.S.Me., P.A.L., G.M., A.D., L.Y., L.F.B., D.Z., P.J.v.d.M., D.S., R.M., G.He., T.Kar., Z.W., T.Li., I.D., J.Z., W.M., L.La., S.W.v.L., J.P.B., A.R.W., A.Bo., T.S.A., L.M.H., E.S., S.Y., I.M.M., L.Ca., H.G.d.H., M.A., U.A., N.A., F.W.A., S.E.B., S.B., A.Ca., Y. Chan, C.C., G.Da., G.E., B.F., M.F.F., F.Ge., M.G., S.E.H., J.J.H., J.H., J.E.H., P.G.H., A.J., Y.K., S.K., R.A.L., B.L., M.Lo., S.J.Loo., Y.L., P.M., A.Ma., C.Men., F.D.M., E.M., M.E.M., A.Mo., A.O., I.Pa., F.P., I.Pr., L.M.R., B.Sa., R.M.S., R.Sa., H.S., W.R.S., C.Sa., C.Ma., B.Se., S.Sh., S.J.Lon., J.A.S., L.S., R.J.S., M.J.S., S.Ta., B.O.T., A.Tog., M.To., N.Ts., J.v.S., S.V., D.Vo., E.B.W., W.W., J.Y., G.Z., N.J.S., R.A.S., A.D.M., C.N.P., S.I.B., N.J.T., A.P.M., S.C.H., H.T., N.Pi., L.P., P.v.d.H., P.Kr., R.Sc., I.B.B., A.Te., C.O., M.C., J.D., J.I.R., N.S., K.E.N., A.Ta., J.C.C., J.S.K. and D.P.S. analysed the data. P.K.J., T.E., H.Ma., N.E., I.Ga., T.N., A.U.J., C.Sc., A.V.Sm., M.C.B. and D.P.S. performed beta-testing of scripts. P.K.J. and T.E. performed the meta-analysis. P.K.J., T.E., O.P. and J.F.W. wrote the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
G.P. is a co-founder of CAVADIS B.V. S.W.v.L. is a former employee of CAVADIS B.V. B.M.P. serves on the Data and Safety Monitoring Board of a clinical trial funded by the LifeVest manufacturer (Zoll Lifecor) and on the Yale Open Data Access Project funded by Johnson & Johnson. N.Po. has received financial support and consultancy fees from several pharmaceutical companies that manufacture either blood-pressure-lowering or lipid-lowering agents or both. P.S. has received research awards from Pfizer. No other authors declared a conflict of interest.
Additional information
Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa, 230-0045, Japan.
Extended data figures and tables
Extended Data Figure 1 Forest plot for cognitive ability (g).
Individual sub-cohort estimates of effect size and the 95% confidence interval are plotted. Sub-cohorts are ordered from top to bottom according to their weight in the meta-analysis, so larger or more homozygous cohorts appear towards the top. The scale of  is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1. This trait was rank-transformed.
is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1. This trait was rank-transformed.
Extended Data Figure 2 Forest plot for educational attainment.
Individual sub-cohort estimates of effect size and the 95% confidence interval are plotted. Sub-cohorts are ordered from top to bottom according to their weight in the meta-analysis, so larger or more homozygous cohorts appear towards the top. The scale of  is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1.
is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1.
Extended Data Figure 3 Forest plot for height.
Individual sub-cohort estimates of effect size and the 95% confidence interval are plotted. Sub-cohorts are ordered from top to bottom according to their weight in the meta-analysis, so larger or more homozygous cohorts appear towards the top. The scale of  is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1.
is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1.
Extended Data Figure 4 Forest plot for forced expiratory lung volume in one second.
Individual sub-cohort estimates of effect size and the 95% confidence interval are plotted. Sub-cohorts are ordered from top to bottom according to their weight in the meta-analysis, so larger or more homozygous cohorts appear towards the top. The scale of  is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1. This trait was rank-transformed.
is in intra-sex standard deviations. The meta-analytical estimate is displayed at the bottom. Sub-cohort names follow the conventions detailed in Supplementary Table 6 and the Supplementary Table 11 legend. Sample sizes, effect sizes and P values for association are given in Table 1. This trait was rank-transformed.
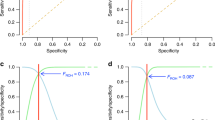
Extended Data Figure 5 Signals of directional dominance are robust to stratification by geography or demographic history or inclusion of educational attainment as covariate.
a, Cohorts are divided by continental biogeographic ancestry (African (15 sub-cohorts), East Asian (5), South and Central Asian (SC Asian; 10), Hispanic (3)), with Europeans being divided into Finns (13), other European isolates (self-declared, 23), and (non-isolated) Europeans (90). Meta-analysis was carried out for all subsets with 2,000 or more samples available. Sample numbers are as follows: cognitive g, Eur isolate, 6,638; European, 44,153; educational attainment, African 4,811; Eur isolate, 8,032; European, 55,549; Finland 9,068; height, African, 21,500; E Asian, 30,011; Eur isolate, 23,116; European, 228,813, Finland, 30,427, Hispanic, 5,469, SC Asian, 13,523; FEV1, African, 6,604, Eur isolate, 4,837, European, 49,223, Finland, 2,340.  is consistent across geography and in both isolates and more cosmopolitan populations. b, Cohorts were divided into high and low ROH strata of equal power and meta-analysis repeated – the effects are consistent across strata for all four traits. The mean SROH for the high and low strata, respectively, are 13.4 and 4.3 Mb for cognitive g; 28.1 and 5.1 Mb for educational attainment; 31.9 and 10.8 Mb for height; and 41.4 and 4.5 Mb for FEV1. c, To assess the potential for socio-economic confounding, where available, educational attainment was included in the regression model (edu) and compared to a model without educational attainment (none) in the same subset of cohorts. The signals reduce slightly when the education covariate is included; the analysis is not possible for educational attainment as a trait. For cognitive g, numbers of subjects are 36,847 and 36,023; for height 131,614 and 120,945; and for FEV1, 15,717 and 15,425, for edu and none, respectively. The numbers differ because of missing individual educational data within cohorts. Plus signs indicate that the phenotype was rank-transformed. Trait units are intra-sex standard deviations and the genomic measure is unpruned SROH. Subset estimates of effect size for FROH and the 95% confidence are plotted.
is consistent across geography and in both isolates and more cosmopolitan populations. b, Cohorts were divided into high and low ROH strata of equal power and meta-analysis repeated – the effects are consistent across strata for all four traits. The mean SROH for the high and low strata, respectively, are 13.4 and 4.3 Mb for cognitive g; 28.1 and 5.1 Mb for educational attainment; 31.9 and 10.8 Mb for height; and 41.4 and 4.5 Mb for FEV1. c, To assess the potential for socio-economic confounding, where available, educational attainment was included in the regression model (edu) and compared to a model without educational attainment (none) in the same subset of cohorts. The signals reduce slightly when the education covariate is included; the analysis is not possible for educational attainment as a trait. For cognitive g, numbers of subjects are 36,847 and 36,023; for height 131,614 and 120,945; and for FEV1, 15,717 and 15,425, for edu and none, respectively. The numbers differ because of missing individual educational data within cohorts. Plus signs indicate that the phenotype was rank-transformed. Trait units are intra-sex standard deviations and the genomic measure is unpruned SROH. Subset estimates of effect size for FROH and the 95% confidence are plotted.
Extended Data Figure 6 Signals of directional dominance are robust to model choice.
Meta-analytical estimates of effect size and standard errors are plotted for various models. Fixed, no mixed modelling was used; gr res, GRAMMAR+ residuals were fitted; hglm, full hierarchical generalized linear mixed model was used. Plus signs indicate that the phenotype was rank-transformed. 15,355 subjects were used for cognitive g, 36,060 for educational attainment, 89,112 for height and 15,262 for FEV1.
Extended Data Figure 7 Correlation in SROH for different genotyping arrays using HapMap populations.
a–c, x and y axes show SROH from 0–30 Mb. ill370, Illumina CNV370; aff6, Affymetrix6; illomni, Illumina OmniExpress. The graphs are shown for the specific PLINK call parameters used. d, Sample numbers per continent are presented in a bar chart. AFR, African; AMR, mixed American; ASN, East Asian; EUR, European; SAN, South Asian. Only samples with SROH below 30 Mb are plotted, to be conservative to the effect of outliers, which have very strongly correlated estimates of SROH (r = 0.96–0.97 for comparisons including such very homozygous individuals). In these plots, the correlation between SROH called by the two arrays, r = 0.93–0.94.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-11. (XLSX 170 kb)
Supplementary Information
This file contains full legends for Supplementary Tables 1-11. (PDF 133 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Joshi, P., Esko, T., Mattsson, H. et al. Directional dominance on stature and cognition in diverse human populations. Nature 523, 459–462 (2015). https://doi.org/10.1038/nature14618
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14618
This article is cited by
-
Associations of genome-wide and regional autozygosity with 96 complex traits in old order Amish
BMC Genomics (2023)
-
From Mendel to quantitative genetics in the genome era: the scientific legacy of W. G. Hill
Nature Genetics (2022)
-
An Amish founder population reveals rare-population genetic determinants of the human lipidome
Communications Biology (2022)
-
Fine-scale population structure and demographic history of British Pakistanis
Nature Communications (2021)
-
Ethnic Identity and Genome Wide Runs of Homozygosity
Behavior Genetics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.