Abstract
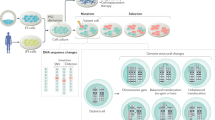
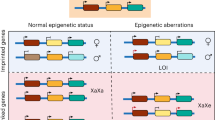
Human pluripotent stem cells hold potential for regenerative medicine, but available cell types have significant limitations. Although embryonic stem cells (ES cells) from in vitro fertilized embryos (IVF ES cells) represent the ‘gold standard’, they are allogeneic to patients. Autologous induced pluripotent stem cells (iPS cells) are prone to epigenetic and transcriptional aberrations. To determine whether such abnormalities are intrinsic to somatic cell reprogramming or secondary to the reprogramming method, genetically matched sets of human IVF ES cells, iPS cells and nuclear transfer ES cells (NT ES cells) derived by somatic cell nuclear transfer (SCNT) were subjected to genome-wide analyses. Both NT ES cells and iPS cells derived from the same somatic cells contained comparable numbers of de novo copy number variations. In contrast, DNA methylation and transcriptome profiles of NT ES cells corresponded closely to those of IVF ES cells, whereas iPS cells differed and retained residual DNA methylation patterns typical of parental somatic cells. Thus, human somatic cells can be faithfully reprogrammed to pluripotency by SCNT and are therefore ideal for cell replacement therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Processed data sets can be downloaded from the NCBI GEO under accession GSE53096 for RNA-seq, SNP array and 450K methylation array, and accession GSE57179 for MethylC-seq data. Analysed MethylC-seq data sets can also be accessed at http://neomorph.salk.edu/SCNT/browser.html.
References
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998)
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007)
Rais, Y. et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502, 65–70 (2013)
Hussein, S. M. et al. Copy number variation and selection during reprogramming to pluripotency. Nature 471, 58–62 (2011)
Laurent, L. C. et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8, 106–118 (2011)
Ruiz, S. et al. Analysis of protein-coding mutations in hiPSCs and their possible role during somatic cell reprogramming. Nature Commun. 4, 1382 (2013)
Nazor, K. L. et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10, 620–634 (2012)
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011)
Ohi, Y. et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nature Cell Biol. 13, 541–549 (2011)
Ruiz, S. et al. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc. Natl Acad. Sci. USA 109, 16196–16201 (2012)
Tachibana, M. et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–1238 (2013)
Lowry, W. E. et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl Acad. Sci. USA 105, 2883–2888 (2008)
Fusaki, N., Ban, H., Nishiyama, A., Saeki, K. & Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn Acad. B 85, 348–362 (2009)
Taylor, R. W. & Turnbull, D. M. Mitochondrial DNA mutations in human disease. Nature Rev. Genet. 6, 389–402 (2005)
Bock, C. et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439–452 (2011)
Suzuki, R. & Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006)
Ziller, M. J. et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 7, e1002389 (2011)
Stelzer, Y. et al. Identification of novel imprinted differentially methylated regions by global analysis of human-parthenogenetic-induced pluripotent stem cells. Stem Cell Rep. 1, 79–89 (2013)
Rugg-Gunn, P. J., Ferguson-Smith, A. C. & Pedersen, R. A. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum. Mol. Genet. 16, R243–R251 (2007)
de Hoon, M. J., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004)
Saldanha, A. J. Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004)
Silva, S. S., Rowntree, R. K., Mekhoubad, S. & Lee, J. T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl Acad. Sci. USA 105, 4820–4825 (2008)
Vallot, C. et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nature Genet. 45, 239–241 (2013)
Newman, A. M. & Cooper, J. B. AutoSOME: a clustering method for identifying gene expression modules without prior knowledge of cluster number. BMC Bioinformatics 11, 117 (2010)
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnol. 28, 495–501 (2010)
Nishino, K. et al. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 7, e1002085 (2011)
Polo, J. M. et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature Biotechnol. 28, 848–855 (2010)
Xie, W. et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148 (2013)
Laurent, L. et al. Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320–331 (2010)
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013)
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009)
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 25, 25–29 (2000)
Gore, A. et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67 (2011)
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 (2010)
Johnson, W. E., Rabinovic, A. & Li, C. Adjusting batch effects in microarray expression data using Empirical Bayes methods. Biostatistics 8, 118–127 (2007)
Price, M. E. et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin 6, 4 (2013)
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, (2011)
Langmead, B. et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Perkins, W., Tygert, M. & Ward, R. Computing the confidence levels for a root-mean-square test of goodness-of-fit. Appl. Math. Comput. 217, 9072–9084 (2011)
Bancroft, T., Du, C. & Nettleton, D. Estimation of false discovery rate using sequential permutation p-values. Biometrics 69, 1–7 (2013)
Schultz, M. D., Schmitz, R. J. & Ecker, J. R. ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28, 583–585 (2012)
Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Huang, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009)
Acknowledgements
The authors acknowledge the OHSU Embryonic Stem Cell Research Oversight Committee and the Institutional Review Board for providing oversight and guidance. We thank oocyte and sperm donors and the Women’s Health Research Unit staff at the Center for Women’s Health, University Fertility Consultants and the Reproductive Endocrinology and Infertility Division in the Department of Obstetrics and Gynecology of Oregon Health and Science University for their support and procurement of human gametes. We are grateful to C. Penedo for microsatellite analysis and W. Sanger and D. Zaleski for karyotyping services. We are also indebted to Y. Li, H. Sritanaudomchai and D. Melguizo Sanchis for their technical support. We thank the staff at the Institute for Genomic Medicine Genomics Facility at UCSD for running the Infinium HumanMethylation450 BeadChips and sequencing of the RNA-seq libraries. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin (http://www.tacc.utexas.edu) and the San Diego Supercomputing Center (through an allocation from the eXtreme Science and Engineering Discovery Environment (XSEDE)) for providing HPC resources that have contributed to the research results reported within this paper. SCNT and iPS cell studies were supported by grants from the Leducq Foundation and OHSU institutional funds. R.M., K.S., R.T. and L.C.L. were supported by the UCSD Department of Reproductive Medicine. Methylome studies were supported by the Salk International Council Chair fund endowment and the Mary K. Chapman Foundation to J.R.E. J.R.E. is an investigator of the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (GMBF3034). A.P. received a fellowship from the Swedish Research Council, Vetenskapsrådet. E.K. was partially funded by a fellowship from the Collins Medical Trust.
Author information
Authors and Affiliations
Contributions
H.M., R.M., L.C.L. and S.M. conceived the study and designed the experiments. P.A., M.S. and N.M.G. coordinated recruitment of gamete donors. P.A. performed ovarian stimulations and oocyte retrievals. M.T., M.S., N.M.G. and S.M. conducted SCNT, IVF and embryo culture experiments. R.T.-H., S.M., M.T., M.S., N.M.G., H.M., A.P., B.D., E.K., A.S. and R.A. derived and cultured IVF ES cells, NT ES cells and iPS cells. S.G. performed teratoma analysis. H.M., M.T. and C.V.D. performed the DNA and RNA extractions, mtDNA amplification refractory mutation system qPCR analyses, and qPCR. R.M., K.S., R.D.T. and L.C.L. performed SNP, DNA methylation and RNA-seq studies and bioinformatic analysis of the data. R.C.O., Y.H., M.D.S., M.H., J.R.N., R.C. and J.R.E. conducted MethylC-seq studies. H.M., R.M., R.C.O., Y.H., J.R.E., L.C.L., D.P.W. and S.M. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Mitochondrial DNA genotyping.
a, Mitochondrial DNA (mtDNA) genotyping by RNA-seq and MethylC-seq. The NT4 line carried a C/T heteroplasmy at position 16092 (open oval) while the other NT ES cell and IVF ES cell lines contained a homoplasmic C allele at this position. b, Chromatographs of single nucleotide polymorphisms (SNPs, arrows) within the human mitochondrial genome indicate that all four NT ES cell lines share a mtDNA sequence with IVF ES cells. Notably, the NT4 line carried a C/T heteroplasmy at position 16092 (double peaks with blue representing C and red representing T in the chromatograph) while other NT ES cell lines and both hESO-7 and hESO-8 contained a homoplasmic C allele. The mtDNA sequence of all iPS cell lines was identical to the parental HDFs. c, mtDNA genotyping by Sanger sequencing demonstrated that all Leigh-iPS cell lines contain a G mutation at mtDNA position 8993 and the Leigh-NT1 line contains oocyte mtDNA with a wild-type T at the same position.
Extended Data Figure 2 Subchromosomal genomic aberrations in IVF ES cells, NT ES cells and iPS cells.
a, The location and type of CNVs for all mapped samples. One-copy deletion regions are shown in red, two-copy deletions are in yellow, duplicated regions (three copies) are in dark blue, and runs of homozygosity (ROHs) are in green. b, The average number of CNVs per stem cell type for IVF ES cells, NT ES cells and iPS cells. Owing to the small sample sizes, no statistically significant differences were found between sample groups. c, Bar graphs displaying the number of InDels by sample. d, Bar graphs showing the average number of InDels found in the iPS cell lines and NT ES cell lines. No statistically significant differences were found between sample groups. Error bars, s.e.m.
Extended Data Figure 3 XIST and XACT expression.
a, Bar graph showing the reads per kb per million reads (RPKMs) of the XIST gene for pluripotent stem cell lines and HDFs. b, Bar graph showing the log transformed normalized read count of the XACT gene for the same samples. Error bars, s.e.m.
Extended Data Figure 4 Genes with aberrant methylation and associated alterations in gene expression.
a, Hypermethylation of iPS-R2 (black line in bar graphs representing the average β-values for methylation level, right side of y axis) and decreased gene expression of POU3F4, SLITRK2 and SLITRK4 (bar graphs representing normalized reads, averaged between replicates, left side of y axis). b, Hypomethylation (black line in bar graphs representing the average β-values for methylation level, right side of y axis) of iPS-R1 (top two graphs and bottom left corner) and iPS-S2 (bottom right corner) correlated with decreased gene expression of DACH2, CHM, RPS6KA6 and TMEM187 (bar graph representing normalized reads, averaged between replicates, left side of y axis).
Extended Data Figure 5 Differential methylation at autosomal non-imprinted loci.
Heat map displaying 1,621 autosomal, non-imprinted CpGs that were differentially methylated among NT ES cells, iPS cells and IVF ES cells (n = 10) (Kruskal–Wallis P-value < 0.01, Δβ > 0.5). CpG probes were clustered into six groups using an unsupervised self-organizing map algorithm24. The line graphs on the right represent an average β-value for each cluster.
Extended Data Figure 6 Methylation of CpG probes.
Box plots representing the β-values for all autosomal non-imprinted probes on the methylation array located within specified genomic regions. The box plots show a general trend of higher methylation levels in iPS cells compared to IVF ES cells. The number of CpGs interrogated in each genomic region is included on the y axis. The box represents the interquartile range (25th to 75th percentile), and the line within the box marking represents the median. The notch in the box represents the 95% confidence interval around the median. The whiskers above and below the box contain 99.3% of the data with outliers represented by circles above and below the whiskers. a, Probes within 2,000 base pairs of the transcription start site (TSS). b, Probes within CpG Islands (CGIs). c, Probes in the 5′ region (0–2 kb upstream of CGI). d, Probes in the 3′ region (0–2 kb downstream of CGI). e, Probes in the 5′ region (2–4 kb upstream of CGI). f, Probes in the 3′ region (2–4 kb downstream of CGI). g, Functional annotation of the mammalian genome (FANTOM 4) promoters with high CpG content. h, FANTOM 4 promoters with low CpG content. i, Probes within enhancers. j, Probes within major histocompatibility complex (MHC) regions. k, Probes within cancer differential methylated regions (CDMRs). l, Probes within reprogramming differentially methylated regions (RDMRs). m, Probes within short interspersed nuclear element (SINE) regions. n, Probes within long interspersed nuclear element (LINE) regions. o, Probes within long terminal repeat (LTR) regions.
Extended Data Figure 7 Non-CG mega DMRs.
a, Heat map of normalized mCH/CH of all 150 non-CG mega DMRs identified by comparing four NT ES cell lines, nine iPS cell lies to five IVF ES cell lines from this study and the previous studies28,29,30. b, An example of non-CG mega DMRs (black bar) ranged from 1,995,000 bp to 4,850,000 bp on chromosome 8. The y axis is normalized mCH/CH, which is defined as the weighted non-CG methylation level minus bisulphite non-conversion and dividing median mCH/CH of 5 kb bin. Scope was extended 200 kb on both sides to show non-CG methylation profile of regions surrounding non-CG mega DMRs. c, A representative non-CG mega DMR (black bar) hypomethylated in both iPS cells and NT ES cells on chromosome 21. d, A representative non-CG mega DMR hypermethylated only in iPS cells on chromosome 10.
Extended Data Figure 8 Expression patterns of genes in non-CG mega DMRs.
a, Number of genes in non-CG mega DMRs identified in each sample. b, Average number of genes falling in non-CG mega DMRs in NT ES cells and iPS cells. c, Histogram of gene expression in iPS cells for the genes located in hypermethylated. d, Hypomethylated non-CG mega DMRs identified in iPS cells. The x axis is the log2 fold change of iPS cell RPKM compared to IVF ES cell RPKM. e, Histogram of gene expression in NT ES cells for the genes located in hypomethylated non-CG mega DMRs. The x axis is the log2 fold change of NT ES cell RPKM compared to IVF ES cell RPKM. NT ES cell (or iPS cell) RPKM was the average of two replicates, while ES cell RPKM was the average of all replicates of hESO-7 and hESO-8.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4. (PDF 238 kb)
Supplementary Table 5
List of CD DMRs (XLSX 231 kb)
Supplementary Table 6
List of non-CG mega-DMRs (XLSX 39 kb)
Supplementary Table 7
GO analysis results for genes within non-CG mega-DMRs (XLSX 25 kb)
Rights and permissions
About this article
Cite this article
Ma, H., Morey, R., O'Neil, R. et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511, 177–183 (2014). https://doi.org/10.1038/nature13551
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13551
This article is cited by
-
A way to wipe a cell’s memory
Nature (2023)
-
Transient naive reprogramming corrects hiPS cells functionally and epigenetically
Nature (2023)
-
BrewerIX enables allelic expression analysis of imprinted and X-linked genes from bulk and single-cell transcriptomes
Communications Biology (2022)
-
Imprinting fidelity in mouse iPSCs depends on sex of donor cell and medium formulation
Nature Communications (2022)
-
hiPSCs for predictive modelling of neurodegenerative diseases: dreaming the possible
Nature Reviews Neurology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.