Abstract
Antibodies are important tools in anatomical pathology and research, but the quality of in situ protein detection by immunohistochemistry greatly depends on the choice of antibodies and the abundance of the targeted proteins. Many antibodies used in scientific research do not meet requirements for specificity and sensitivity. Accordingly, methods that improve antibody performance and produce quantitative data can greatly advance both scientific investigations and clinical diagnostics based on protein expression and in situ localization. We demonstrate here protocols for antibody labeling that allow specific protein detection in tissues via bright-field in situ proximity ligation assays, where each protein molecule must be recognized by two antibodies. We further demonstrate that single polyclonal antibodies or purified serum preparations can be used for these dual recognition assays. The requirement for protein recognition by pairs of antibody conjugates can significantly improve specificity of protein detection over single-binder assays.
Similar content being viewed by others
Main
Immunohistochemistry has a central role in pathological examination. The technique has remained essentially the same since the early 1940s,1 and it is increasingly used for diagnostic phenotyping of cancers. Immunohistochemistry plays a pivotal role in cancer biomarker discovery and validation, helping to characterize the distribution of specific proteins in affected tissues.2, 3, 4 The biological relevance of signals generated using immunohistochemistry greatly depends on the quality of the antibodies used, necessitating careful validation of reagents.5 Despite extensive antibody research the availability of specific antibodies for in situ protein detection remains limiting for progress.6 Probably a majority of both poly- and monoclonal antibodies can bind either homologous or non-homologous epitopes on proteins other than those against which they have been raised.7 This can result in troubling cross-reactivity, especially when the cross-reactive proteins are abundant relative to those being targeted for analysis.8 Optimization of experimental conditions by varying antibody concentrations or procedures for blocking can improve performance, but may still not permit specific detection.9
It is well known that antibodies that cross-react with several target proteins may nonetheless be useful in assays where detection depends on simultaneous binding by pairs of antibodies to distinct epitopes of the same target protein molecule.10 This dual recognition mechanism is commonly used in sandwich immunoassays for detecting proteins in solution, and it is also an element of in situ proximity ligation assay (PLA).6, 11, 12, 13 In in situ proximity ligation assay two oligonucleotide-modified antibodies, brought in close proximity by binding to pairs of epitopes present on the same or on two interacting target proteins, guide circularization of two other oligonucleotides, thereby permitting localized rolling-circle amplification for enhanced detection. The single-stranded rolling-circle amplification products are visualized using labeled detection oligonucleotides complementary to a repeated motif in the rolling-circle amplification products.14 The requirement for recognition by pairs of antibodies in in situ proximity ligation assay has so far mainly been used for analyzing protein-protein interactions or proteins with post-translational modifications,15, 16 but the dual recognition aspect can also help improve accuracy for in situ detection of individual proteins in cells and tissues, or in western blots.17 A key requirement for in situ proximity ligation assay is the efficient conjugation of oligonucleotides to antibodies.18 Current conjugation protocols typically require high antibody concentrations and purification steps to remove unwanted material from the antibody preparation, complicating high-throughput applications.19, 20, 21
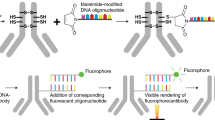
The main aims of this work were to develop simplified procedures for antibody functionalization with DNA strands for in situ proximity ligation assay analyses of protein expression, and to evaluate the reagents for specific visualization of proteins in tissues. Affinity-purified polyclonal rabbit antibodies (www.proteinatlas.org) and immunoglobulin preparations purified from rabbit serum using protein A/G were tested for performance in immunohistochemistry and in situ proximity ligation assay. We related the results, displayed as protein expression in tissue sections, to the levels of transcripts from the corresponding genes in those same tissues, in order to assess technical characteristics of the assays such as specificity and limits of detection. We optimized a method for high-throughput conjugation of antibodies with DNA strands for use in in situ proximity ligation assay experiments. The direct conjugation to primary antibodies circumvents the need to use antibodies derived from different animal species in order to apply these with oligonucleotide-conjugated secondary antibodies that are species-selective for each of the two primary antibodies (Figure 1a). We furthermore demonstrate three approaches to construct reagents for in situ proximity ligation assay using (a) pairs of antibody preparations from the same species, raised against different parts of the same target protein (Figure 1b), (b) total protein A/G-purified IgG, or (c) affinity-purified polyclonal antibody preparation (Figure 1c). We illustrate that in situ proximity ligation assay provides a convenient and general means to significantly improve the specificity of protein detection in situ compared to standard immunohistochemistry.
Approaches for protein detection using in situ proximity ligation assays. (a) Two primary antibodies from different species bind the same target protein molecule and are recognized by oligonucleotide-conjugated secondary antibodies, selective for each of the two primary antibodies. The attached DNA strands are brought in proximity, allowing them to template ligation of two subsequently added oligonucleotides to form a DNA circle. The DNA circle is then amplified by localized rolling-circle amplification, and the amplification product is visualized by hybridization with complementary HRP-labeled oligonucleotides. (b) In situ proximity ligation assay using a pair of oligonucleotide-modified primary antibodies from the same species, raised against different parts of the target protein, or (c) using a pair of DNA conjugates generated from a single polyclonal antibody preparation.
Materials and methods
Antibodies
The primary antibodies were obtained from the Human Protein Atlas Program (www.proteinatlas.org) and from Antibody Portal at National Cancer Institute (http://antibodies.cancer.gov/), and had been affinity purified by binding to the immunizing peptide, except when noted, and initially validated according to standard immunohistochemistry protocols.3 The antibodies, their dilutions for immunohistochemistry experiments, and information about the antigens are listed in Supplementary Table 1.
Antibody Purification
Antibodies were purified using Zeba spin desalting 7 K MWCO columns (89882 columns, Thermo Fisher Scientific). Thirty microliter of each antibody was applied to the center of the compacted resin bed, followed by the addition of 15 μl of PBS to the top of the resin to ensure maximal protein recovery. The columns were centrifuged at 1500 × g for 2 min to collect the desalted sample.
Serum Purification
Crude sera from immunized rabbits were purified with a Melon Gel IgG Purification Kit (Piercenet) according to the manufacturer’s protocol. The sera were diluted 1:10 in 1 × Purification Buffer and added to the column packed with Melon Gel resin. After 5 min incubation buffer was added and fractions were collected. The concentration of the recovered IgG was measured by NanoDrop (Thermo Fisher Scientific).
Conjugation Using the Duolink Probemaker Kit
Conjugation with the Probemaker kit (Sigma-Aldrich) was performed essentially according to the manufacturer’s protocol, except that affinity-purified antibodies at concentrations ranging from 0.1 to 0.3 μg/μl were supplemented with rabbit gamma immunoglobulin (011-000-002, Jackson Immunoresearch) to a final total antibody concentration of 1 μg/μl in PBS to increase the protein concentration to that recommended by the manufacturer. Antibodies were divided in two aliquots of 20 μl each for conjugation with anti-mouse PLUS and anti-rabbit MINUS oligonucleotides. Two microliter of Conjugation Buffer was added to each aliquot, and the solution was transferred to a vial of lyophilized, reactive oligonucleotide and incubated at room temperature overnight. Addition of 2 μl of Stop Reagent was followed by incubation for 30 min at room temperature. Next 24 μl of Storage Solution was added and the conjugated antibody was stored at 4°C, ready for use. For preparation of total IgG in situ proximity ligation assay probes IgG samples were divided in two aliquots (1 μg/μl in 20 μl), and conjugated with the two oligonucleotides as described above.
Conjugation via DBCO
Ten microliters of antigen-specific antibodies was supplemented with total rabbit gamma immunoglobulin (011-000-002, Jackson Immunoresearch) to a final concentration of 2 μg/μl in PBS. The antibodies were activated for 30 min at RT with 0.4 μM the bifunctional crosslinker dibenzylcyclooctyne-NHS ester (DBCO), dissolved in dimethyl sulfoxide (DMSO). After buffer exchange with Zeba Spin Desalting Columns (molecular weight cut-off 7000; Thermo Fisher Scientific), removing excess crosslinker, the activated antibodies were incubated with a 4-fold molar excess of either 5′ or 3′ azide-modified oligonucleotides (IDT) at 4 °C overnight for coupling via click chemistry.22 The conjugates were then stored in PBS with 0.02% NaN3 at 4 °C with no further purification.
Sample Preparation for Immunohistochemistry and In Situ Proximity Ligation Assay
Tissue microarray or cell array sections of formalin fixed human healthy tissue were mounted onto Super Frost/Plus slides (Menzel-Glaser), and baked at 60 °C for 45 min. After deparaffinization in xylene and hydration in graded alcohols, endogenous peroxidase activity was blocked in 0.3% hydrogen peroxide. Antigen retrieval was performed by pressure boiling in a Decloaking Chamber (Biocare Medical) in Target Retrieval Solution (S169, Dako) at 125 °C for 4 min.
Immunohistochemistry
The slides were stained with primary antibodies (Supplementary Table 1) in an XL ST5010 (Leica Microsystems GmbH), followed by detection via Polymer Quanto and the horseradish peroxidase (HRP)-UltraVision kit (Thermo Fischer Scientific), as described previously.23 The slides were mounted with Pertex (Histolab AB), and scanned using an Aperio ScanScope XT (Aperio Technologies).
In situ Proximity Ligation Assay
For in situ proximity ligation assay, the tissue microarray and cell microarray slides were blocked in Duolink blocking solution (Sigma-Aldrich) for 1 h. Alternatively, for specific antibody conjugates prepared with added nonimmune IgG, the Duolink blocking solution was supplemented with rabbit IgG (011-000-002, Jackson Immunoresearch) at a concentration of 2 μg/μl in PBS and incubated overnight. Next, the slides were incubated overnight at 4 °C with a combination of two primary antibodies, either oligonucleotide-conjugated or not, diluted in Duolink antibody diluent (Sigma-Aldrich). For control experiments only one primary antibody or conjugate was used. Following incubation with primary mouse and rabbit antibodies lacking conjugated oligonucleotides, anti-mouse PLUS and anti-rabbit MINUS (Sigma-Aldrich) in situ proximity ligation assay secondary probes were applied to the tissues, and incubated at 37 °C for 2 h. The slides were then washed in Tris-buffered saline with Tween (50 mM Tris pH 8.4, 150 mM NaCl, 0.05% Tween 20). Both slides reacted with unconjugated and conjugated primary antibodies were then ready for incubation with the ligation solution for 30 min at 37 °C. This solution contained a pair of oligonucleotides capable of forming a DNA circle, templated by the oligonucleotides conjugated to primary or secondary antibodies and catalyzed by a DNA ligase. This was followed by incubation with an Amplification Solution (Sigma-Aldrich) for an additional 90 min at 37 °C, producing local DNA amplification products via rolling-circle amplification at sites of binding by pairs of primary antibodies. After washing in Tris-buffered saline with Tween the amplification products were visualized by hybridizing HRP-conjugated DNA probes (5′-CAGTGAATGCGAGTCCGTCT-3′, Biomers) diluted in SSC buffer containing 0.05% Tween 20, 0.4 μg/μl BSA and 0.01 μg/μl polyadenine (Sigma), followed by incubation with DAB substrate (Thermo Fisher Scientific) for 1 min. Slides were counterstained with hematoxylin and eosin (Histolab), and then mounted with Pertex (Histolab). To investigate the possibility of unspecific binding of conjugates to the tissue and false signal formation the tissue microarrays were incubated with the single conjugates (HPA000956+, HPA002564−, HPA051783+, HPA051783−, HPA010512+, HPA010512−, HPA003425+, HPA003425-) that was followed by incubation with proximity ligation assay reagents (Supplementary Figure 1).
Gene Expression Analysis
Total RNA from three tissue sections was extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions, and the RNA quality was analyzed with an Agilent 2100 Bioanalyzer system with the RNA 6000 Nano LabChip Kit (Agilent Biotechnologies). The mRNA strands were fragmented using Fragmentation Buffer (Illumina) and the templates were used to construct cDNA libraries using a TruSeq RNA Sample Prep Kit (Illumina). Gene expression was assessed by deep sequencing of cDNA on Illumina HiSeq HiSeq2000/2500 system (Illumina) for paired-end reads with a read length of 2 × 100 bases. The mRNA levels were quantified, normalized, and scored as fragments per kilobase of exon model per million mapped reads using gene models from Ensembl build 69 with Cufflinks 2.0.2. The gene expression analysis was performed as a part of the Human Protein Atlas project (www.proteinatlas.org),3, 24 and the primary RNA sequencing data has also been submitted to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-1733.
Ethical Approval
All human samples used in this study were waived by the Uppsala Ethical Review Board [Reference # 2002–577, 2005–338 and 2007–159 (protein) and # 2011-473 (RNA)].
Results
In order to broadly implement dual recognition in situ protein detection assays with oligonucleotide-modified reagents, it is important to have simple, reliable means of conjugating DNA strands to the antibodies. We therefore began by investigating the effects of click chemistry-based conjugation of low amounts of antibodies in the presence of total nonimmune rabbit IgG. We compared results in standard immunohistochemistry and in situ proximity ligation assay experiments on paraffin fixed formalin embedded tissue sections and paraffin-embedded cells, using unmodified or DNA-modified antibodies. We used two rabbit antibodies exhibiting good specificity for their targets, HPA000956 and HPA002564. The two rabbit antisera had been generated against two distinct, recombinant protein fragments of 118 and 149 amino acids, respectively, from the APEX-1 protein (APEX nuclease (multifunctional DNA repair enzyme 1), and previously shown to give specific staining by immunohistochemistry (www.proteinatlas.org). First, we applied the antibodies for immunohistochemistry on tissue microarrays of healthy tissues. Both antibodies produced closely similar immunohistochemistry staining patterns in sections from several types of tissue with a predominantly nuclear expression pattern, and weaker cytoplasmic staining in tissues from ovary, uterus, and placenta (Figure 2). Next, we modified both antibodies with the bifunctional DBCO reagent and conjugated them to azide-modified oligonucleotides. During conjugation the specific antibodies were supplemented with total nonimmune rabbit IgG to a final concentration of 2 mg/ml to avoid the need to concentrate the specific antibodies to the required concentration for the conjugation reaction. We performed in situ proximity ligation assay with the pairs of antibody-oligonucleotide conjugates on serial tissue sections. The protein expression levels as reported by both immunohistochemistry and in situ proximity ligation assay correlated to the mRNA levels in the various tissues and cells, supporting the staining specificity and demonstrating that the presence of total rabbit IgG during conjugation does not negatively impact the performance of the conjugates (Supplementary Figure 2, Supplementary Figure 3). In addition, we performed control proximity ligation assay experiments omitting one conjugate at the time. For proximity ligation assay signal to be generated two binding events bringing probes in close proximity are necessary, therefore any signal generated in such an experiment is considered to be unspecific. In our study following conjugates: HPA000956+, HPA002564−, HPA051783+, HPA051783−, HPA010512+, HPA010512−, HPA003425+, HPA003425 haven’t generated any unspecific signals when incubated with tissue unaccompanied (Supplementary Figure 1). The most similar patterns were observed in ovarian tissue. In the uterus, no cytoplasmic signal was seen in in situ proximity ligation assay-stained tissue whereas in placenta the cytoplasmic signal was enhanced by in situ proximity ligation assay compared to HPA002564. In paraffin-embedded cells we showed the change in staining intensities in two different cell lines (U-87 and U-937) whose mRNA levels differed by 4-fold (Supplementary Figure 2).
Detection of the APEX-1 protein in human ovary, uterus and placenta tissue by immunohistochemistry using affinity-purified HPA000956 or HPA002564 rabbit antibodies, and in situ proximity ligation assay (PLA) with DNA-modified HPA000956 and HPA002564 antibodies, conjugated along with added nonimmune rabbit IgG to achieve a sufficient protein concentration for convenient conjugation. Similar patterns of staining are seen with all reagents. The APEX-1 mRNA expression in each organ is shown as fragments per kilobase of exon model per million mapped reads (FPKM).
We next tested a commercially available conjugation kit for preparation of in situ proximity ligation assay probes (Olink Bioscience). Due to the low concentrations of the affinity-purified antibodies available to us (0.1 to 0.3 μg/μl), the manufacturer’s protocol was again modified by addition of nonimmune rabbit IgG to reach the required total protein concentration. We compared immunohistochemistry with the use of conjugates of pairs of polyclonal antibodies raised against different segments of the same protein with results from single polyclonal antibody preparations that were divided in two aliquots and conjugated with distinct oligonucleotides. These two strategies to prepare reagents for in situ proximity ligation assay were applied in detection reactions for the well-known heart-specific protein biomarker troponin T type 2 (TNNT2) and calcium/calmodulin-dependent protein kinase II CAMK2A-D family of proteins. Each of the two preparations of antibodies directed against TNNT2 used in our study (HPA015774 and HPA017888) have been previously shown to be heart specific (www.proteinatlas.org)25 and were good candidates to investigate different approaches to prepare detection reagents. Positive immunohistochemistry staining generated with HPA015774 and HPA017888 was only seen in heart tissue, while other tissues such as cerebral cortex remained unstained. The corresponding fragments per kilobase of exon model per million mapped reads values for mRNA expression in heart and cerebral cortex confirmed the specificity of these staining reactions. Prior to in situ proximity ligation assay analysis, tissue samples were incubated with total IgG to block any unspecific binding of the oligonucleotide-conjugated nonimmune IgG. In situ proximity ligation assay staining using three different combinations of affinity reagents (HPA015774+/HPA017888-, HPA015774+/HPA015774− and HPA017888+/HPA017888−) all resulted in similar staining patterns of varying intensities, depending on the combination used. A slight increase of background for in situ proximity ligation assay in sections from cerebral cortex was seen compared to regular immunohistochemistry suggesting possible cross-reactivity of the TNNT2 antibodies with TNNT1 (Figure 3). We conclude that the in situ proximity ligation assay signals were quite comparable to those for immunohistochemistry, demonstrating that the nonimmune IgG used during conjugation did not affect the performance of the reagents.
Comparison of in situ proximity ligation assay and immunohistochemistry for detection of the proteins TNNT2 (HPA015774 and HPA017888 antibodies) and CAMK2A-D (HPA051783 and HPA051783 antibodies). In situ proximity ligation assay was performed using a pair of PLUS (+) and MINUS (−) DNA conjugates generated with Duolink Probemaker from single polyclonal antibody preparation or from two rabbits immunized with distinct peptides. The mRNA levels are presented as fragments per kilobase of exon model per million mapped reads (FPKM).
We further validated the conjugation technique with antibodies generated against two short epitopes common for the CAMK2A-D protein. The two CAMK2A-D antibody preparations, directed against two non-overlapping peptides of 26 and 33 amino acids (Supplementary Table 1) from the CAMK2A-D protein (HPA053973 and HPA051783), respectively, generated staining of different intensities by immunohistochemistry. HPA051783 immunohistochemistry staining was equally strong in cerebral cortex and adrenal gland tissue, although mRNA levels of CAMK2A in the brain tissue were considerably higher compared to those of adrenal gland tissue. The HPA053973 antibody generated weaker signals in both tissues with good correlation to mRNA levels. A stronger staining was observed in neurons. We evaluated the performance of different combinations of antibody conjugates (HPA053973+/HPA051783−, HPA053973+/HPA053973−, HPA051783+/HPA051783−) in in situ proximity ligation assay, and found that all resulted in specific signals that correlated well with mRNA levels (Figure 3). HPA053973 staining of neurons was emphasized by using combinations of both HPA053973 and HPA051783 antibodies, despite the unspecific immunohistochemistry staining pattern of the HPA051783 antibody alone.
Immunohistochemistry using polyclonal rabbit antisera is often hampered by problems of unspecific background staining. Having established protocols for antibodies functionalization for in situ proximity ligation assay using highly specific antibodies as shown in Figures 2 and 3, we next selected antibodies whose specificity had not been previously verified. For these experiments, we used antibodies that were raised against peptides from the CLPS (Colipase) and TFF1 (Trefoil factor 1) proteins to investigate in situ protein detection by in situ proximity ligation assay for specific and background staining. Both CLPS and TFF1 encode small secreted proteins with tissue-specific expression at the mRNA level in pancreas (CLPS) and stomach (TFF1), respectively. The CLPS antibody (HPA010512) was generated by immunization with a recombinant protein fragment of 83 amino acids, while the TFF1 antibody (HPA003425) was raised against a protein fragment of 53 amino acids (Supplementary Table 1). Each of the two antibodies were divided into two aliquots and separately conjugated with the two oligonucleotides required for in situ proximity ligation assay (+ and −) so that signals could only be generated by pairs of antibodies binding two epitopes on the same or neighboring protein molecules. Upon conjugation, the two modified antibody preparations were combined and applied for in situ proximity ligation assay. The results were compared with those obtained by immunohistochemistry, and with mRNA levels in the corresponding tissues. The immunohistochemistry signal generated with polyclonal CLPS antibodies was strongest in pancreas, correlating with the prominent mRNA levels. However, signals were also present at relatively high levels in endometrial stroma and spleen where little or no mRNA encoding these proteins was detected. By contrast, when the polyclonal antibodies directed against a peptide from the CLPS protein were split in two aliquots and conjugated to PLUS and MINUS oligonucleotides for dual recognition via in situ proximity ligation assay signals for the CLPS protein were found almost exclusively in pancreatic tissue sections, with minimal or no signals in all other investigated tissues, in excellent correlation with mRNA levels (Supplementary Figure 4). The TFF1 protein was detected at high levels in stomach using both immunohistochemistry and in situ proximity ligation assay in good agreement with mRNA levels, and both methods detected TFF1 in gastric glands, although signal intensities and cellular patterns varied. The in situ proximity ligation assay signals were mostly localized at the inner parts of the glands, while immunohistochemistry staining was present throughout the gland structures. TFF1 protein was detected at moderate to high levels in prostate luminal cells by immunohistochemistry, although the mRNA expression level was very low. A weak immunohistochemistry staining was present in testis where no transcripts encoding TFF1 were found. Unlike the results from immunohistochemistry no TFF1 protein was detected by in situ proximity ligation assay in either prostate or testis tissue sections (Figure 4), consistent with the results of RNA seq.
Immunohistochemistry and in situ proximity ligation assay (PLA) of human tissue by using HPA010512 and HPA003425 antibodies raised against CLPS and TFF1 proteins, respectively. For in situ proximity ligation assay each antibody preparation was divided in two aliquots and modified with PLUS and MINUS oligonucleotides with the addition of nonimmune total rabbit IgG. The mRNA expression for each protein is shown as fragments per kilobase of exon model per million mapped reads (FPKM).
To further investigate means of simplifying the preparation of reagents for in situ proximity ligation assay we used two portions of total IgG purified from serum of an immunized rabbit using protein A and G, and compared this to affinity-purified material, for construction of PLUS and MINUS antibody-oligonucleotide conjugates using Duolink ProbeMaker. No nonimmune IgG was added to these conjugation reactions. We applied the assays for detecting two target proteins: APEX-1 and S100A4 (S100 calcium-binding protein A4). For preparation of in situ proximity ligation assay probes, total rabbit IgG was divided in two aliquots and conjugated to DNA oligonucleotides, and the conjugates were combined and applied for in situ proximity ligation assay. We validated this simple and convenient method for preparing reagents without affinity purification to results obtained from standard in situ proximity ligation assay. The standard assay involves pairs of specific primary antibodies, a mouse monoclonal and affinity-purified rabbit polyclonal antibodies, detected using oligonucleotide-conjugated, species specific secondary antibodies (Figure 1a). The results were also compared to those obtained by immunohistochemistry staining with either total rabbit serum or the corresponding affinity-purified antibodies. The rabbit antibodies used for validation were affinity purified from the same serum sample that was also used for preparation of total IgG and oligonucleotide conjugation. The immunohistochemistry staining developed with both affinity-purified APEX-1 antibodies and anti-APEX-1 IgG was strong in spermatocytes and spermatogonia of testis. Both reagents also generated signals in testicular stroma that were more robust when total IgG was used in immunohistochemistry. The stromal staining was considerably weaker when in situ proximity ligation assay with mouse and rabbit antibodies was applied and absent when conjugates were constructed from two portions of total IgG. In the ovary immunohistochemistry using the affinity-purified antibodies and protein A/G-purified serum all performed similarly as in testis, as did in situ proximity ligation assay. The mRNA levels correlated well with all staining intensities except for that of total IgG immunohistochemistry in testis, which was stronger than expected. Similar observations were made when S100A4 serum was used on cerebral cortex. No specific staining of neurons could be seen due to high background levels when total IgG was used for immunohistochemistry. However, in situ proximity ligation assay with total IgG conjugates generated very specific signals in neurons by in situ proximity ligation assay. The proximity ligation assay signals generated with purified IgG were similar to staining generated with primary mouse and rabbit antibodies combined with anti-mouse PLUS and anti-rabbit MINUS secondary probes. All reagents and assays generated comparable signals in spleen, and consistent with mRNA expression profiles from the same tissues. The results demonstrated that both in situ proximity ligation assay protocols performed equally well for detecting targeted proteins, and that the specificity of detection was greatly improved when total IgG was used for in situ proximity ligation assay compared to results from immunohistochemistry (Figure 5).
Comparison of in situ proximity ligation assay (PLA) and immunohistochemistry for detection of APEX-1 and S100A4 proteins. IHC was performed using either affinity-purified antibodies (HPA002564 or HPA007973, specific for APEX-1 and S100A4, respectively) or protein A/G-purified IgG from the same rabbit sera. In situ proximity ligation assay was performed with pairs of primary mouse and rabbit antibodies towards targeted proteins (HPA002564/CAB047307 for APEX-1 and HPA007973/CAB027388 for detecting S100A4), or using total rabbit IgG divided into two aliquots and conjugated separately to DNA oligonucleotides. The staining intensities are correlated with mRNA levels shown as fragments per kilobase of exon model per million mapped reads (FPKM). In situ proximity ligation assay was performed with pairs of primary mouse and rabbit antibodies directed towards the targeted proteins. In panel a APEX was targeted using antibodies HPA002564 and CAB047307). In panel b S100A4 was targeted with antibodies HPA007973 and CAB027388.
Discussion
Immunohistochemistry is widely used for in situ protein detection in tissue samples. It has a central role in clinical pathology for detecting the presence, localization, and abundance of specific proteins in tissue sections. The technique serves to establish diagnoses, classify neoplasms, and provide therapeutic or prognostic information. Although simple in wconcept, immunohistochemistry usually requires considerable optimization, as many factors may influence the outcome. The selection of suitable poly- or monoclonal antibodies is one of the most important factors influencing immunohistochemistry and the diagnostic outcome. Problematic immunohistochemistry staining is frequently a consequence of low detection sensitivity or insufficiently specific target molecule detection, both strongly influenced by antibody quality. Proper antibody titration is necessary to select the dilution that offers the greatest signal/background ratio, but it can be difficult to know what the correct signal is and what constitutes background.26
Despite the importance for diagnostic decisions, interpretation of protein expression via immunohistochemistry is done in a qualitative and subjective manner, with limited opportunities for quantification. Accordingly, methods to achieve higher detection specificity could improve clinical diagnostics by allowing a greater understanding of protein expression and disease mechanisms. In this regard, the in situ proximity ligation assay technique offers a number of valuable characteristics, as follows: (i) The requirement for simultaneous recognition by pairs of antibodies rather than by single antibodies can greatly improve specificity of in situ detection assays, and (ii) it also allows detection of interacting or post-translationally modified proteins. (iii) The localized amplification via rolling-circle amplification upon successful target detection serves to improve sensitivity of detection. (iv) Rolling-circle amplification also results in discrete detection signals that can be objectively recorded for digital quantification of staining in situ, either by manual inspection or via dedicated software.
In our previous work we established a protocol for HRP-based bright-field in situ proximity ligation assay for protein detection in formalin fixed paraffin-embedded tissue, allowing in situ proximity ligation assay to be applied with bright-field microscopy in routine histopathology.17 Combinations of mouse monoclonal and rabbit polyclonal antibodies were applied along with commercially available secondary anti-rabbit and anti-mouse antibodies with attached oligonucleotides such that colocation of pairs of primary mouse and rabbit antibodies resulted in specific visualization of proteins in situ.
In the present work we demonstrate robust, convenient, and specific means to detect proteins via in situ proximity ligation assay by directly conjugating total or affinity-purified polyclonal antibodies from immunized rabbits for in situ protein detection with enhanced specificity through dual recognition. We developed a strategy for simplified and efficient conjugation of oligonucleotides to small amounts of antibodies, and we investigated the effect upon specificity of detection by dual recognition in situ proximity ligation assay. By supplementing protein solutions with nonimmune IgG to final concentrations optimal for chemical modification we could use minimal concentrations of specific antibodies for conjugation. Antibody–DNA conjugates were successfully generated from affinity-purified antibodies at concentrations of 0.1 mg/ml. The approach was applied using two different conjugation methods. The first one, commercially available from Sigma-Aldrich, employed a convenient all-in-one conjugation kit. We also tested a less costly, alternative method, where DBCO-modified antibodies were reacted with oligonucleotides having terminal azide residues. Also, this second method proved suitable for high-throughput experiments, but it required some optimization and hands-on experience. Using both these protocols, several polyclonal antibodies were modified with oligonucleotides, allowing us to analyze their performance in in situ proximity ligation assay.
We demonstrate herein the potential of in situ proximity ligation assay to improve specificity and to reduce or altogether avoid nonspecific background staining. We applied the antibody conjugates for in situ proximity ligation assay using three different approaches; (1) affinity-purified polyclonal rabbit antibodies from two rabbits immunized with distinct peptides from the same protein were used for conjugation, or (2) affinity-purified antibodies from one rabbit were divided in two aliquots before conjugation, or, finally, (3) oligonucleotides were conjugated to two aliquots of total IgG from one immunized rabbit. For every experiment, we evaluated the correlation between data obtained using in situ proximity ligation assay with those obtained by immunohistochemistry, and with expression of the corresponding genes as mRNA in the same human tissues.
It can be difficult to distinguish between specific immunohistochemistry signals and background due to antibody cross-reactivity. Nonspecific background can occur when antibodies recognize sufficiently similar and relatively abundant epitopes on proteins other than those targeted for analysis. This is common for closely related proteins, such as G-coupled receptor proteins,27 but it may also occur for unrelated proteins. Depending on the similarities of epitopes and the relative concentrations of targeted and cross-reactive proteins it may not be possible to avoid such background even by careful antibody titration. We show herein that in situ proximity ligation assay offers greater selectivity for the correct target, without the need for as extensive antibody titration as needed for immunohistochemistry. In situ proximity ligation assay revealed subtle differences in expression among cells and different cell compartments that could not be observed either through immunohistochemistry or by global mRNA analysis of the tissues.
Among polyclonal antibodies from an immunized rabbit only some 0.1%–5% (depending on titer) typically exhibit affinity for the antigen used for immunization, motivating affinity purification of antibodies.28 We demonstrate herein that in situ proximity ligation assay, where any protein must be detected by two antibody molecules, can override the need for affinity purification, simplifying the preparation of suitable detection reagents. Antisera raised against whole protein regularly encompass antibodies with affinity for several non-overlapping epitopes. We show that in situ proximity ligation assay could be used with two aliquots of antibodies raised against a peptide of only 26 amino acids suggesting that this short motif nonetheless includes two distinct epitopes that can be recognized by pairs of antibodies without interference. The generality of this observation will have to be investigated further.
In conclusion, in situ proximity ligation assay offers improved performance in protein detection, both for sensitivity and specificity, compared to the more traditional immunohistochemistry technique. Accordingly, in situ proximity ligation assay using poly- or monoclonal antibodies can be a cost-effective method in research and for clinical screening of patient material. We present herein several protocols that simplify the direct labeling of antibodies with oligonucleotides, as required in these analyses. We next plan to extend our studies to greater numbers of targets and tissues to establish the generality of our conclusions, and to further investigate the prognostic and predictive value of in situ proximity ligation assay in pathology.
References
Coons AH . The development of immunohistochemistry. Ann N Y Acad Sci 1971;177:5–9.
Brennan DJ, O'Connor DP, Rexhepaj E et al. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat Rev Cancer 2010;10:605–617.
Uhlen M, Fagerberg L, Hallstrom BM et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419.
Uhlen M, Oksvold P, Fagerberg L et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248–1250.
O'Hurley G, Sjostedt E, Rahman A et al. Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol Oncol 2014;8:783–798.
Buchwalow I, Samoilova V, Boecker W et al. Non-specific binding of antibodies in immunohistochemistry: fallacies and facts. Sci Rep 2011;1:28.
Anagnostou VK, Welsh AW, Giltnane JM et al. Analytic variability in immunohistochemistry biomarker studies. Cancer Epidemiol Biomarkers Prev 2010;19:982–991.
Fritschy JM . Is my antibody-staining specific? How to deal with pitfalls of immunohistochemistry. Eur J Neurosci 2008;28:2365–2370.
Voskuil J . Commercial antibodies and their validation. F1000Res 2014;3:232.
Fredriksson S, Gullberg M, Jarvius J et al. Protein detection using proximity-dependent DNA ligationassays. Nat Biotechnol 2002;20:473–477.
Blokzijl A, Nong R, Darmanis S et al. Protein biomarker validation via proximity ligation assays. Biochim Biophys Acta 2014;1844:933–939.
Kamali-Moghaddam M, Pettersson FE, Wu D et al. Sensitive detection of Abeta protofibrils by proximity ligation—relevance for Alzheimer's disease. BMC Neurosci 2010;11:124.
Vashist SK, Marion Schneider E, Lam E et al. One-step antibody immobilization-based rapid and highly-sensitive sandwich ELISA procedure for potential in vitro diagnostics. Sci Rep 2014;4:4407.
Soderberg O, Gullberg M, Jarvius M et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 2006;3:995–1000.
Grannas K, Arngarden L, Lonn P et al. Crosstalk between Hippo and TGFbeta: subcellular localization of YAP/TAZ/Smad complexes. J Mol Biol 2015;427:3407–3415.
Zieba A, Pardali K, Soderberg O et al. Intercellular variation in signaling through the TGF-beta pathway and its relation to cell density and cell cycle phase. Mol Cell Proteomics 2012;11:M111 013482.
Zieba A, Wahlby C, Hjelm F et al. Bright-field microscopy visualization of proteins and protein complexes by in situ proximity ligation with peroxidase detection. Clin Chem 2010;56:99–110.
Kozlov IA, Melnyk PC, Stromsborg KE et al. Efficient strategies for the conjugation of oligonucleotides to antibodies enabling highly sensitive protein detection. Biopolymers 2004;73:621–630.
Takeda S, Tsukiji S, Nagamune T . Site-specific conjugation of oligonucleotides to the C-terminus of recombinant protein by expressed protein ligation. Bioorg Med Chem Lett 2004;14:2407–2410.
Wu AM, Senter PD . Arming antibodies: prospects and challenges for immunoconjugates. Nat Biotechnol 2005;23:1137–1146.
Agaton C, Falk R, Hoiden Guthenberg I et al. Selective enrichment of monospecific polyclonal antibodies for antibody-based proteomics efforts. J Chromatogr A 2004;1043:33–40.
Khatwani SL, Kang JS, Mullen DG et al. Covalent protein-oligonucleotide conjugates by copper-free click reaction. Bioorg Med Chem 2012;20:4532–4539.
Kampf C, Olsson I, Ryberg U et al. Production of tissue microarrays, immunohistochemistry staining and digitalization within the human protein atlas. J Vis Exp 2012;63:e3620.
Fagerberg L, Hallstrom BM, Oksvold P et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13:397–406.
Lindskog C, Linne J, Fagerberg L et al. The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody-based profiling. BMC Genomics 2015;16:475.
Baker M . Reproducibility crisis: blame it on the antibodies. Nature 2015;521:274–276.
Michel MC, Wieland T, Tsujimoto G . How reliable are G-protein-coupled receptor antibodies? Naunyn Schmiedebergs Arch Pharmacol 2009;379:385–388.
Bradbury A, Pluckthun A . Reproducibility: standardize antibodies used in research. Nature 2015;518:27–29.
Acknowledgements
This work was supported by the Knut and Alice Wallenberg Foundation (#2008:0143), the European Community's 7th Framework Program (FP7/2007–2013) under grant agreement n° 222635 (AffinityProteome) 241481 (Affinomics), The Swedish Research Council, Swedish Governmental Agency for Innovation Systems, IngaBritt and Arne Lundberg Foundation, the European Research Council under the European Union's Seventh Framework Programme (FP/2007–2013) / ERC Grant Agreement n. 294409 (ProteinSeq), and Uppsala University. UL holds stock in Olink, having rights to the in situ proximity ligation assay technology. We would also like to thank Tara Hiltke at the National Cancer Institute for providing mAbs for in situ proximity ligation assay experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Zieba, A., Ponten, F., Uhlén, M. et al. In situ protein detection with enhanced specificity using DNA-conjugated antibodies and proximity ligation. Mod Pathol 31, 253–263 (2018). https://doi.org/10.1038/modpathol.2017.102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.102
This article is cited by
-
Improved efficiency of in situ protein analysis by proximity ligation using UnFold probes
Scientific Reports (2018)