Abstract
Immunohistochemical analysis of cyclin-dependent kinase inhibitor 1C (CDKN1C, p57, Kip2) expression and molecular genotyping accurately classify hydatidiform moles into complete and partial types and distinguish these from non-molar specimens. Characteristics of a prospective series of all potentially molar specimens encountered in a large gynecologic pathology practice are summarized. Initially, all specimens were subjected to both analyses; this was later modified to triage cases for genotyping based on p57 results: p57-negative cases diagnosed as complete hydatidiform moles without genotyping; all p57-positive cases genotyped. Of the 678 cases, 645 were definitively classified as complete hydatidiform mole (201), partial hydatidiform mole (158), non-molar (272), and androgenetic/biparental mosaic (14); 33 were unsatisfactory, complex, or problematic. Of the 201 complete hydatidiform moles, 104 were p57-negative androgenetic and an additional 95 were p57-negative (no genotyping), 1 was p57-positive (retained maternal chromosome 11) androgenetic, and 1 was p57-non-reactive androgenetic; 90 (85%) of the 106 genotyped complete hydatidiform moles were monospermic and 16 were dispermic. Of the 158 partial hydatidiform moles, 155 were diandric triploid, with 154 p57-positive, 1 p57-negative (loss of maternal chromosome 11), and 1 p57-non-reactive; 3 were triandric tetraploid, with 2 p57-positive and 1 p57-negative (loss of maternal chromosome 11). Of 155 diandric triploid partial hydatidiform moles, 153 (99%) were dispermic and 2 were monospermic. Of the 272 non-molar specimens, 259 were p57-positive biparental diploid, 5 were p57-positive digynic triploid, 2 were p57-negative biparental diploid (no morphological features of biparental hydatidiform mole), and 6 were p57-non-reactive biparental diploid. Of the 14 androgenetic/biparental mosaics with discordant p57 expression, 6 were uniformly mosaic and 8 had a p57-negative androgenetic molar component. p57 expression is highly correlated with genotyping, serves as a reliable marker for diagnosis of complete hydatidiform moles, and identifies androgenetic cell lines in mosaic conceptions. Cases with aberrant and discordant p57 expression can be correctly classified by genotyping.
Similar content being viewed by others
Main
Accurate subclassification of molar specimens into complete hydatidiform mole and partial hydatidiform mole and distinction of these from non-molar specimens are important for clinical management and for accurate assessment of the risk of persistent gestational trophoblastic disease. The risk of persistent gestational trophoblastic disease for complete hydatidiform moles (15–20%) is significantly higher than for partial hydatidiform moles (0.2–4%).1, 2, 3, 4 Despite the lower risk associated with partial hydatidiform moles, metastatic gestational trophoblastic disease and trophoblastic tumors coexistent with or subsequent to a diagnosis of partial hydatidiform mole have been reported.5, 6, 7, 8 In addition, distinction of partial hydatidiform moles from non-molar specimens is important for appropriate clinical management, as a diagnosis of partial hydatidiform mole generates follow-up with serum beta human chorionic gonadotropin (HCG) levels and contraception, which would be unnecessary following a diagnosis of a non-molar specimen and undesirable for infertility patients. However, previous studies have demonstrated that diagnosis of hydatidiform moles based on morphology alone, even by experienced pathologists with specialized training, is subject to interobserver variability and therefore suboptimal diagnostic reproducibility.9, 10, 11, 12, 13, 14, 15 A number of studies have demonstrated the value of ancillary techniques, including immunohistochemical analysis of cyclin-dependent kinase inhibitor 1C (CDKN1C/p57/Kip2, the protein product of the CDKN1C imprinted gene located at chromosome 11p15.5; referred to henceforth as p57) expression16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 and molecular genotyping via PCR amplification of short tandem repeat loci,23, 25, 28, 29, 30, 31 for improving the diagnosis of hydatidiform moles. Genotyping is particularly valuable because it allows for specific distinction of complete hydatidiform moles, partial hydatidiform moles, and non-molar specimens from one another due to their unique genetics. Complete hydatidiform moles are purely androgenetic and usually diploid (two paternal chromosome complements without a maternal chromosome complement),32 with most resulting from fertilization of an egg by a single sperm that duplicates (monospermy)33, 34 and a minority via fertilization by two sperms (dispermy),35 with loss of the maternal chromosome complement. In contrast, partial hydatidiform moles are characterized by diandric triploidy (two paternal and one maternal chromosome complements), with most arising by dispermy.36, 37, 38 Non-molar specimens are usually characterized by biparental diploidy (one paternal and one maternal chromosome complements). Some non-molar specimens are digynic triploid conceptions (two maternal and one paternal chromosome complements) but do not exhibit molar features,39 with the exception of rare examples having the morphology and immunophenotype (p57-negative) of complete hydatidiform moles occurring in patients with familial recurrent hydatidiform mole associated with mutations in NLRP7 (NALP7) or KHDC3L (C6orf221).40 In addition, some non-molar specimens with cytogenetic abnormalities such as trisomy can simulate partial hydatidiform moles, causing problems in diagnostic reproducibility.41, 42
In 2007, we began a prospective analysis of all potentially molar products of conception specimens encountered on the Gynecologic Pathology Service of The Johns Hopkins Hospital, Baltimore, MD using immunohistochemical analysis of p57 expression and molecular genotyping with short tandem repeat markers.25, 31, 43 The current study summarizes the results of nearly 6 years of analysis, providing a summary of the characteristics of a large series of hydatidiform moles and non-molar specimens assessed by these techniques as well as the assessment of the performance of these methods in clinical practice.
Materials and methods
All cases encountered on the Gynecologic Pathology Service of The Johns Hopkins Hospital, Baltimore, MD for which there was any clinical or pathological concern for a molar specimen were prospectively analyzed according to the diagnostic algorithm shown in Figure 1. From July 2007 through March 2013, 678 cases were analyzed, with 86% representing consultation cases and 14% representing routine in-house cases. Initially, all specimens were subjected to both p57 immunohistochemical analysis and molecular genotyping with short tandem repeat markers. After sufficient experience with the reliability of p57 analysis for identifying androgenetic complete hydatidiform moles, this process was later modified to triage cases for genotyping based on p57 results. Thereafter, in the setting of appropriate morphology features, p57-negative cases were diagnosed as complete hydatidiform moles without genotyping and all p57-positive cases continued to be subjected to genotyping.25, 31 In addition, any cases for which p57 immunohistochemical analysis was unsatisfactory were genotyped. In certain uncommon situations involving cases that had morphology and p57 results in our laboratory consistent with a diagnosis of complete hydatidiform mole but outside ancillary testing suggesting a different diagnosis, genotyping was performed to confirm our results. These included cases for which outside laboratory testing reported a DNA index/ploidy result consistent with triploidy and cases in which some focal or apparently discordant p57 expression was noted on a p57 immunostain performed at an outside laboratory but was not seen on the p57 immunostain performed in our laboratory.
Algorithmic approach to diagnosis of hydatidiform moles. Potentially molar specimens were evaluated according to this process. Initially, all specimens were subjected to both p57 immunohistochemical analysis and molecular genotyping with short tandem repeat markers. Cases were diagnosed as complete hydatidiform moles when genotyping demonstrated androgenetic diploidy, as partial hydatidiform moles when genotyping demonstrated diandric triploidy, and as non-molar specimens when genotyping demonstrated biparental diploidy. This process was later modified to triage cases for genotyping based on p57 results, with p57-negative cases being diagnosed as complete hydatidiform moles without genotyping and all p57-positive cases being subjected to genotyping.
Immunohistochemical analysis of p57 expression was performed on 4-μ formalin-fixed paraffin-embedded sections, which were stained using a Ventana BenchMark XT automated immunostaining system (Tucson, AZ, USA) with mouse monoclonal antibodies against p57 protein (Neomarkers, Fremont, CA, USA) from a ready-to-use preparation without dilution. To interpret immunohistochemical stains for p57, the presence or absence of nuclear positivity was assessed in villous stromal cells, cytotrophoblast, intermediate trophoblast, and maternal decidua. The p57 immunostain was interpreted as ‘negative’ and satisfactory when maternal decidua and/or intermediate trophoblastic cells exhibited nuclear expression of p57 (serving as internal positive control in all the cases, including complete hydatidiform moles) but villous stromal cells and cytotrophoblast were either entirely negative or demonstrated only limited expression (nuclear staining in <10% of these cell types). This negative result was then interpreted as consistent with a diagnosis of a complete hydatidiform mole, provided the morphology of the specimen was appropriate. The p57 immunostain was interpreted as ‘positive’ when the extent of staining in these cell types was extensive or diffuse. Nuclear expression in villous stromal cells and cytotrophoblast in the focally positive range (≥10% but <50%) has been considered an ‘equivocal’ result in our previous studies, but when using a binary system of ‘positive’ and ‘negative’ for final interpretation these equivocal results are considered positive based on our experiences, indicating that these equivocal cases have never been confirmed as complete hydatidiform moles per genotyping. The p57 expression pattern was interpreted as ‘discordant’ when there was any combination/admixture of negative and positive results for villous stromal cells and cytotrophoblast within individual villi, including positive staining in cytotrophoblast and negative staining in villous stromal cells, or vice versa. Discordant p57 expression characterizes androgenetic/biparental mosaic/chimeric conceptions, with discordant expression in different cell types based on the presence or absence of maternal genetic material in those particular cells (described in detail in our recent study).44 The p57 expression pattern was further interpreted as ‘divergent’ when two populations of villi, each with different morphologies, exhibited two different staining patterns, eg, a twin gestation comprised of a p57-negative androgenetic diploid complete hydatidiform mole and a p57-positive biparental diploid non-molar specimen or a mosaic specimen comprised of a p57-discordant non-molar specimen component and a p57-negative complete hydatidiform mole component.
Molecular genotyping was performed using unstained 10-μ formalin-fixed paraffin-embedded tissue sections prepared using PCR precautions. A serial H&E-stained 4-μ section was used to identify well-separated areas of maternal decidua and villous tissue. An area of each tissue type was circled with a marking pen on the H&E slide. An unstained slide was superimposed on top of the marked H&E slide and Pinpoint solution (Zymo Research, Orange, CA) was applied to the areas of interest. The tissue was removed from the slide and digested per the manufacturer’s protocol. DNA was further purified using QIAamp DNA spin columns (Qiagen, Valencia, CA). PCR amplification of nine short tandem repeat loci from eight different chromosomes (chromosomes 2,3,4,5,7,11,12,13) and the amelogenin locus (for XY determination) was performed, with thermal cycling conditions and capillary electrophoresis carried out according to the manufacturer’s instructions (AmpFlSTR Profiler kit; Applied Biosystems; Foster City, CA). In certain selected situations (see results), an expanded analysis with PCR amplification of 15 short tandem repeat loci from 13 different chromosomes (chromosomes 2,3,4,5,7,8,11,12,13,16,18,19,21) and the amelogenin locus (for XY determination) was performed (AmpFlSTR Identifiler kit; Applied Biosystems). The PCR conditions were 95 °C for 11 min followed by 28 cycles of 94 °C for 1 min, 59 °C for 1 min, and 72 °C for 1 min, followed by a final extension at 60 °C for 45 min. After amplification, 1 μl of multiplex PCR product was mixed with 9 μl of deionized formamide/GeneScan 500 [ROX] size standard (Applied Biosystems). Samples were denatured at 95 °C for 2 min and placed on ice for at least 1 min before analysis (ABI3100 and ABI 3130xl Genetic Analyzers, Applied Biosystems). Capillary electrophoresis data were analyzed per criteria in our previously published validation study.31 Both the maternal and villous tissues were analyzed to identify alleles at each locus. The villous tissue was further analyzed as follows. Every allele from the villous tissue was identified as being maternal, definitively non-maternal (assumed paternal), or equivocal (unknown whether maternal or paternal due to shared alleles) in origin. For each locus with two alleles identified, the allelic ratio was calculated by dividing the peak height of the longer allele by the peak height of the shorter allele. Allelic ratios between 0.61 and 1.17 were considered to be consistent with diploidy. Allelic ratios between 0.33 and 0.60 or 1.5 and 2.0 were considered to be consistent with triploidy. Loci with three alleles identified in a 1:1:1 ratio were also considered consistent with triploidy. The origin of triploidy was determined when possible based on combined evaluation of allele ratios and source for those alleles with sufficient polymorphism. Each locus was scored as being consistent with diploidy or triploidy or not informative. At least two informative loci were required for final interpretation. The complete hydatidiform moles subjected to genotyping were diagnosed as such by the finding of purely androgenetic alleles. The vast majority are characterized by androgenetic diploidy (two sets of paternal chromosome complements, without a maternal chromosome complement) but a small subset can have androgenetic tetraploidy (genotyping does not specifically distinguish diploid examples from tetraploid examples as peak heights do not indicate actual DNA content). Partial hydatidiform moles were diagnosed by the finding of diandric triploidy (three sets of chromosome complements, two paternal in origin and one maternal in origin (paternal:maternal allele ratio of 2:1)) or triandric tetraploidy (four sets of chromosome complements, with one maternal in origin and three paternal in origin (paternal:maternal allele ratio of 3:1)). Non-molar specimens, including those with abnormal villous morphology related to other non-molar genetic alterations such as trisomy, were diagnosed as such when the genotyping demonstrated biparental diploidy (one maternal and one paternal chromosome complement) and also when digynic triploidy (two maternal and one paternal chromosome complements) was identified, as these latter specimens do not have molar features and are not considered a form of hydatidiform mole. Mosaic/chimeric conceptions were diagnosed by the finding of p57-discordant staining patterns combined with genotyping demonstrating an excess of androgenetic alleles with variable paternal:maternal allele ratios ≥2:1, indicating admixtures of androgenetic and biparental cell lines within individual villi (details provided in a recent study).44
Results
The study included a total of 678 cases. Diagnostic categorization of cases and age distribution are summarized in Table 1. The mean and median ages for all diagnostic categories were similar (27–32 years); the small subset of invasive complete hydatidiform moles had older mean and median ages (39/42 years). Age ranges were similar for partial hydatidiform moles, non-molar specimens, and mosaic cases (13–46 years combined) but different for complete hydatidiform moles, with the age range for complete hydatidiform moles extending beyond age 46 years (up to 55 years). Thus, all patients aged >46 years had complete hydatidiform moles (n=22; mean/median=50/49.5 years, with 11 aged ≥50 years).
All the 678 cases were subjected to p57 immunohistochemical analysis and 583 were subjected to genotyping per the algorithm (Figure 1). Detailed results of immunohistochemical analysis of p57 expression and genotyping are summarized in Tables 2 and 3. Representative examples are illustrated in Figures 2, 3, 4, with corresponding genotyping data presented in Figures 5, 6, 7, 8, 9. In all, 665 of the 678 cases (98%) subjected to p57 immunohistochemical analysis had satisfactory p57 results (Table 2), with 13 (2%) unsatisfactory (non-reactive) or suboptimal (limited weak/equivocal expression in the setting of suboptimal internal positive control) due to degenerative changes and/or technical factors. Also, 558 of the 583 cases (96%) subjected to genotyping yielded satisfactory results (Table 3). Of the 652 cases (96%) for which successful diagnostic results were obtained by either immunohistochemical analysis and/or genotyping, 645 (99%) were definitively classified. These 645 cases were classified as complete hydatidiform mole in 201, as partial hydatidiform mole in 158, as non-molar specimen in 272, and as androgenetic/biparental mosaic in 14. The remaining 33 cases included 7 with complex genotypes that were not readily classifiable, 1 problematic case (discussed in more detail below), and 25 with unsuccessful genotyping analysis. In these 25 cases, unsuccessful genotyping was attributable to insufficient villi, villi being too intimately admixed with decidua for successful microdissection of pure tissue components, or a lack of decidua that precluded definitive interpretation even if the villous tissue was successfully analyzed (required for comparison of villous and maternal DNA patterns). Thus, of the 652 cases that were successfully analyzed, 359 (55%) were typical hydatidiform moles (complete hydatidiform moles and partial hydatidiform moles combined); inclusion of an additional 8 androgenetic/biparental mosaics with a molar (androgenetic complete hydatidiform mole) component raised the total molar cases to 367 (56%).
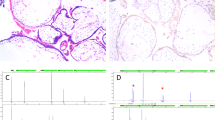
(a, b). Early complete hydatidiform mole displays bulbous villous growth with trophoblastic hyperplasia on villous tips, cellular stroma with small canalicular vessels, and karyorrhectic nuclear debris (a). Villous stromal cells and cytotrophoblast are negative for p57 (b; intermediate trophoblastic cells serve as internal positive control). Genotyping demonstrated androgenetic diploidy (see Figure 5). (c, d). Complete hydatidiform mole demonstrates enlarged villi with notable circumferential trophoblastic hyperplasia (c). Villous stromal cells and cytotrophoblast are diffusely positive for p57 (d), usually arguing against a diagnosis of a complete hydatidiform mole; however, genotyping demonstrated androgenetic diploidy with trisomy 11 due to a retained maternal copy (see Figure 6). (e–g). Partial hydatidiform mole is characterized by hydropic irregularly shaped villi with scalloped contours (e), trophoblastic inclusions (f), and mild circumferential trophoblastic hyperplasia (g). Villous stromal cells and cytotrophoblast are diffusely positive for p57 (h). Genotyping demonstrated diandric triploidy (see Figure 7).
(a, b). Partial hydatidiform mole displays an immature bulbous villous structure with trophoblastic hyperplasia on villous tips and slightly cellular myxoid stroma with small canalicular vessels, simulating an early complete hydatidiform mole (a). Villous stromal cells and cytotrophoblast are diffusely positive for p57 (b), arguing against a diagnosis of an early complete hydatidiform mole. Genotyping demonstrated diandric triploidy, confirming a diagnosis of partial hydatidiform mole (data not shown). (c, d). Partial hydatidiform mole displays an immature villous structure with mild trophoblastic hyperplasia on villous tips and cellular myxoid stroma with karyorrhectic nuclear debris, simulating an early complete hydatidiform mole (c). Villous stromal cells and cytotrophoblast are negative for p57 (d), usually supporting a diagnosis of an early complete hydatidiform mole; however, genotyping demonstrated diandric triploidy with androgenetic diploidy at chromosome 11 due to loss of the maternal copy (see Figure 8). (e–g). Non-molar specimens with abnormal villous morphology demonstrate irregular villi with scalloped villous contours (e), mild trophoblastic hyperplasia (f) and trophoblastic inclusions (g), simulating a partial hydatidiform mole. Villous stromal cells and cytotrophoblast are positive for p57 in both the examples (f, h), consistent with either a non-molar specimen or a partial hydatidiform mole. Genotyping demonstrated biparental diploidy for both the examples, with other markers identifying trisomies as a potential explanation for the abnormal villous morphology (see Figure 9).
(a, b). Non-molar androgenetic/biparental mosaic specimen is comprised of variably sized hydropic villi, with some having cisterns and trophoblastic inclusions and some stromal hypercellularity (a). Villous stromal cells and cytotrophoblast demonstrate a discordant pattern of p57 expression, with uniformly positive cytotrophoblastic cells and negative stromal cells (b). Genotyping demonstrated an excess of paternal alleles with paternal:maternal allele ratios >2:1 (data not shown—see Lewis et al44 for examples). (c–h). Androgenetic/biparental molar mosaic specimen is characterized by two discrete components, a molar component with typical features of a complete hydatidiform mole (c (villi in the left portion have trophoblastic hyperplasia), e) and a mosaic component (c (villi in the right portion lack trophoblastic hyperplasia), g). In some examples, an individual villous structure can display a hybrid of molar and non-molar mosaic features (d; trophoblastic hyperplasia present along upper aspect but lacking along lower aspect). Molar component has trophoblastic hyperplasia (e) and is negative for p57 (f), whereas non-molar mosaic component lacks trophoblastic hyperplasia (g) and displays a discordant pattern of p57 expression (h). Genotyping demonstrated androgenetic diploidy in the molar component and an excess of paternal alleles with paternal:maternal allele ratios >2:1 in the non-molar mosaic component (data not shown—see Lewis et al44 for examples).
Genotyping result for early complete hydatidiform mole in Figure 2a. All loci demonstrate only paternal alleles (no maternal alleles) consistent with androgenetic diploidy (monospermic XX) (blue arrows indicate paternal alleles in the villous tissue that are not present in the decidual tissue).
Genotyping result for p57-positive complete hydatidiform mole in Figure 2c. Four loci (amelogenin, D13S317, D7S820, and CSF1PO) demonstrate only paternal alleles (no maternal alleles) consistent with androgenetic diploidy (dispermic XY). Three chromosome 11 markers (THO1, D11S1981, and D11S2001) demonstrate trisomy of chromosome 11, with two loci (D11S1981, and D11S2001) demonstrating that one copy is maternal in origin (THO1 demonstrates trisomy but is uninformative with respect to parental origin due to allele sharing) (blue arrows indicate paternal alleles in the villous tissue that are not present in the decidual tissue; pink arrows indicate maternal alleles; purple arrows indicate shared alleles of uncertain parental origin).
Genotyping result for partial hydatidiform mole in Figures 2e–g. Two loci (amelogenin and CSF1PO) demonstrate diandric triploidy (dispermic XXY). Three additional loci (VWA, FGA, and D7S820) also demonstrate triploidy but are uninformative with respect to parental origin due to allele sharing (blue arrows indicate paternal alleles in the villous tissue that are not present in the decidual tissue; pink arrows indicate maternal alleles; purple arrows indicate shared alleles of uncertain parental origin).
Genotyping result for p57-negative partial hydatidiform mole in Figure 3c. Two loci (amelogenin and FGA) demonstrate diandric triploidy (dispermic XXY); TPOX demonstrates triploidy but is uninformative with respect to parental origin due to allele sharing. Two chromosome 11 markers (THO1 and D11S2001) demonstrate androgenetic diploidy, indicating loss of the maternal copy of chromosome 11 (blue arrows indicate paternal alleles in the villous tissue that are not present in the decidual tissue; pink arrows indicate maternal alleles; purple arrows indicate shared alleles of uncertain parental origin).
Genotyping result for the non-molar example in Figure 3e. All loci demonstrate biparental diploidy (blue arrows indicate paternal alleles in the villous tissue that are not present in the decidual tissue; pink arrows indicate maternal alleles).
All the 201 complete hydatidiform moles had p57 analysis and 106 were genotyped. Of the 201 complete hydatidiform moles, 104 were p57-negative androgenetic (Figures 2a and b, Figure 5) and an additional 95 were p57-negative (no genotyping), 1 was p57-positive androgenetic with aberrant p57 expression attributable to a retained maternal copy of chromosome 11 (location of the p57 gene) (Figures 2c, d, and 6), and 1 was p57-non-reactive androgenetic (technically unsatisfactory immunostain). Of the 106 genotyped androgenetic complete hydatidiform moles, 90 (85%) were monospermic (XX) and 16 (15%) were dispermic (13 XY, 3 XX) (Table 4). The complete hydatidiform moles included four multiple gestations comprised of a p57-negative androgenetic diploid complete hydatidiform mole and p57-positive biparental diploid non-molar specimen (three twins, one quintuplet). There were seven invasive complete hydatidiform moles encountered in hysterectomy specimens. Of these, five were monospermic XX and two were dispermic XY, with the monospermic examples confirmed as such by analysis with the expanded panel of markers (Identifiler kit; Table 5). Three of the seven invasive complete hydatidiform moles had atypical trophoblastic proliferations morphologically consistent with choriocarcinoma. Of these, two were monospermic (XX) and one was dispermic (XY); one of each of these types had lung nodules consistent with metastatic gestational trophoblastic disease.
All the 158 partial hydatidiform moles had both p57 analysis and genotyping. Of the 158 partial hydatidiform moles, 155 were diandric triploid, with 154 p57-positive (Figures 2e–h, Figures 3a and b, Figure 7), 1 p57-negative with loss of p57 expression attributable to loss of the maternal copy of chromosome 11 (Figures 3c and d, Figure 8), and 1 p57-non-reactive (technically unsatisfactory); 3 partial hydatidiform moles were triandric tetraploid, with 2 p57-positive and 1 p57-negative with loss of p57 expression attributable to loss of the maternal copy of chromosome 11. Based on genotyping with the standard panel of markers (Profiler kit), the 155 diandric triploid partial hydatidiform moles were dispermic in 150 (97%) and monospermic in 5 (3%). Using the expanded panel of markers (Identifiler kit) to further evaluate the five monospermic examples, three of these five were found to be dispermic. Thus, based on the combined analysis, 153 (99%) were dispermic (83 XXY, 57 XXX, 13 XYY) and 2 (1%) were monospermic (both XXX) (Table 6). The three triandric tetraploid partial hydatidiform moles were at least dispermic (two XXYY, one XXXY).
All the 272 non-molar specimens had both p57 analysis and genotyping. Of the 272 non-molar specimens, 259 were p57-positive biparental diploid (Figures 3e–h and 9), 5 were p57-positive digynic triploid, 2 were p57-negative biparental diploid (uncertain etiology for loss of p57 expression as there were no morphological features of complete hydatidiform mole to suggest familial biparental hydatidiform mole), and 6 were p57-non-reactive (related to degenerative changes) biparental diploid. The digynic triploid cases were also analyzed with the expanded panel of markers (Identifiler kit) to enhance the strength of the data for establishing that there was no evidence of diandry (with an increased number of loci demonstrating triploidy with only shared alleles in double dosage (that is, no evidence of paternal alleles in double dosage), the chances of diandric triploidy become extremely small). Among the non-molar specimens, 11 trisomies were identified using the 9-marker panel, including 9 single trisomies involving chromosomes 2, 3, 4, 5 (2 cases), 7 (2 cases), and 13 (2 cases), and 2 double trisomies involving chromosome 4 and 7 and 7 and 13. It is worth noting that the nine-marker panel does not include markers for several of the chromosomes most commonly affected by trisomy in first trimester spontaneous abortions, including chromosomes 16, 18, 21, and 22; so many of those non-molar specimens with abnormal villous morphology potentially attributable to trisomies are not identified by this analysis. In 2 non-molar specimens subjected to analysis with the expanded 15-marker panel for academic interest, 2 additional trisomies were identified, including 1 single trisomy involving chromosome 21 and 1 double trisomy involving chromosomes 16 and 21. Also for academic interest, further investigation of the non-molar specimen with double trisomy involving chromosomes 7 and 13 (which was initially interpreted as a partial hydatidiform mole by morphology) via single-nucleotide polymorphism array analysis identified triple trisomy of paternal origin involving chromosomes 7, 13, and 20.42
There were 21 cases with more complex genotypes than those of typical molar and non-molar specimens. Of these, 14 were classifiable as forms of androgenetic/biparental mosaic specimens (11 of these are described in detail in our previous study).44 These included six uniformly mosaic specimens characterized by villi that were hydropic but lacked trophoblastic hyperplasia and had discordant p57 expression in villous stromal cells and cytotrophoblast throughout (Figures 4a and b). Genotyping demonstrated an excess of androgenetic alleles with variable paternal:maternal allele ratios ≥2:1, indicating admixtures of androgenetic and biparental cell lines within individual villi (see previous study for genotyping example).44 There were eight androgenetic/biparental mosaic specimens that had two distinct components within each case. One component was characterized by p57-discordant villi lacking trophoblastic hyperplasia and the other component was characterized by p57-negative villi with trophoblastic hyperplasia morphologically typical of a complete hydatidiform mole (Figures 4c–h). Genotyping demonstrated an excess of androgenetic alleles with variable paternal:maternal allele ratios ≥2:1 in the non-molar mosaic component and androgenetic diploidy in the molar component (when of sufficient size to allow for analysis of a pure region) (see previous study for genotyping example).44 The remaining seven cases had problematic complex genotyping data and were not readily classifiable as any of the established diagnostic categories described above.
One particularly problematic case was encountered. It was a very early abortus characterized by immature chorionic villi with circumferential sheet-like trophoblastic hyperplasia, suggesting the possibility of an early complete hydatidiform mole; degenerative changes were also present, with some villi appearing necrotic. The p57 analysis was suboptimal and equivocal, with most villi appearing negative but some having very focal faint expression in the setting of focal yet suboptimal internal positive control expression in intermediate trophoblastic cells; this suboptimal result was also obtained on repetition of the assay and was likely related to the degenerative changes noted in the specimen. Genotyping demonstrated biparental diploidy, including informative results for the marker on chromosome 11 (location of the p57 gene). The combined findings were interpreted as equivocal for an early complete hydatidiform mole of familial biparental type versus an early abortus with trophoblastic hyperplasia, with a recommendation for some follow-up of the serum HCG level. Given the rarity of familial biparental hydatidiform moles, the potentially negative but suboptimal/equivocal p57 result, and the lack of available history of recurrent hydatidiform moles or ancillary genetic testing to identify any of the known mutations associated with this disorder, we could not establish a definitive diagnosis of a familial biparental hydatidiform mole for this case.
Discussion
The current study of a large prospective series of potentially molar products of conception specimens analyzed with p57 immunohistochemistry and short tandem repeat genotyping provides a comprehensive summary of the characteristics of molar and non-molar specimens assessed with these techniques. Both p57 analysis and genotyping can be successfully performed in a high proportion of cases (96–98%), even when specimens are derived from a variety of outside laboratories (86% consultation cases) with likely variable specimen fixation times/processing methods from laboratory to laboratory. These techniques thereby allow for definitive classification of a high proportion of those cases with successful ancillary testing results (99%).
In the current study, a substantial proportion of the cases (86%) was derived from our gynecologic pathology consultation service, so we acknowledge some degree of bias in this study set. Despite this, the set included a wide morphological spectrum of cases, ranging from typical complete hydatidiform moles and partial hydatidiform moles with characteristic diagnostic features (including complete hydatidiform moles with prominent trophoblastic hyperplasia and invasive complete hydatidiform moles in hysterectomy specimens) to typical early complete hydatidiform moles, to subtle forms of complete hydatidiform moles and partial hydatidiform moles, as well as non-molar specimens with a spectrum of appearances. Interestingly, this prospectively collected set has had since its beginning both a substantial number of hydatidiform moles (55% of the total cases in the current study set) and a greater proportion of complete hydatidiform moles than partial hydatidiform moles (complete hydatidiform mole:partial hydatidiform mole ratio=1.3 in the current study set). This has been a surprise to us, given that diagnosis of partial hydatidiform moles and non-molar specimens is more problematic than complete hydatidiform moles and one would expect partial hydatidiform moles to outnumber complete hydatidiform moles in a set of consultation cases. Although early forms are expected to constitute a notable subset of complete hydatidiform moles (given that a substantial number of hydatidiform moles are encountered at relatively early gestational ages since the introduction of routine imaging studies early in pregnancy), they did not constitute the exclusive form of complete hydatidiform mole represented in this set (as judged by subjective morphological assessment, as genotyping is identical for all forms of complete hydatidiform moles). Thus, we believe the study set is comprised of a wide spectrum of cases, including a sufficient number of complete hydatidiform moles, suggesting that the database is not unduly biased.
One interesting finding in this series was the observation that all cases encountered in women aged >46 years were complete hydatidiform moles (that is, none were partial hydatidiform moles or non-molar specimens). This distinct predilection toward complete hydatidiform mole over partial hydatidiform mole in women over a particular age has been observed in previous studies.45, 46 We did not have specific data on the menopausal status of the older women in our study but of the 22 patients aged >46 years who had complete hydatidiform moles (age range=47–55), 8 were aged ≥52 years and 11 were aged ≥50 years, indicating that many of these women were likely perimenopausal and that some were probably postmenopausal. Although uncommon, examples of complete hydatidiform moles in postmenopausal women ranging in age from 51 to 61 years have been reported.47, 48
This large series demonstrates that complete hydatidiform moles are virtually always p57-negative, with only rare examples (0.5%) displaying aberrant (positive) p57 expression, attributable to retention of the maternal copy of chromosome 11.49 Complete hydatidiform moles are androgenetic conceptions by definition and the vast majority are monospermic (85%). Partial hydatidiform moles are almost always p57-positive, with only rare examples (1.3%) exhibiting aberrant (negative) p57 results, attributable to the loss of the maternal copy of chromosome 11.50 Partial hydatidiform moles are also almost always diandric triploid conceptions (98%), with nearly all being dispermic (99%). The rarity of monospermic partial hydatidiform moles observed in our study is similar to results reported in a recent study.51 Rare partial hydatidiform moles (2%) are triandric tetraploid.52 The rare examples of both complete hydatidiform moles and partial hydatidiform moles with aberrant p57 expression can be correctly classified by genotyping. These aberrant cases present an interesting problem for classification of hydatidiform moles. The p57-negative partial hydatidiform moles would have been interpreted as complete hydatidiform moles based on the p57 result if genotyping had not been performed on these cases. The distinction of a partial hydatidiform mole from a complete hydatidiform mole is important, because the risk of persistent gestational trophoblastic disease differs for these entities, being significantly higher for complete hydatidiform moles (15–20%) than for partial hydatidiform moles (0.2–4%).1, 2, 3, 4 Potential ‘over-diagnosis’ as a complete hydatidiform mole is without significant clinical consequence, as appropriate follow-up with serum HCG levels would occur, perhaps for a longer time than if diagnosed as a partial hydatidiform mole, but there would be little harm in opting for that more conservative (safe) approach other than possibly requiring contraception for a somewhat longer period. However, ‘under-diagnosis’ of a complete hydatidiform mole with aberrant retained p57 expression as a partial hydatidiform mole using only p57 immunohistochemistry without molecular genotyping would underestimate the potential risk of persistent gestational trophoblastic disease and could lead to insufficient clinical evaluation and follow-up.
Our study establishes that immunohistochemical analysis of p57 expression is highly correlated with genotyping results. Only 3 of the 359 hydatidiform moles (0.8%) had aberrant p57 expression (that is, contrary to the expected result for the diagnostic category). In conjunction with previous results demonstrating the high reproducibility (kappa value of 0.9) of interpretation of p57 immunostains,14, 15 the findings demonstrate that p57 immunohistochemistry is extremely reliable for diagnosis of complete hydatidiform moles. Thus, the algorithmic approach for diagnosis of hydatidiform moles we advocate that uses p57 results to triage cases for genotyping (Figure 1) provides a highly reliable method for accurate diagnosis of complete hydatidiform moles in routine practice using a single immunohistochemical stain, with very little risk of misclassification of complete hydatidiform moles. Consequently, genotyping of complete hydatidiform moles is not necessary in routine practice and can be reserved for problematic cases, such as when p57 immunostaining is suboptimal or unsatisfactory or when morphology and p57 results appear discrepant (as in the p57-positive complete hydatidiform mole shown in Figures 2c and d). One exceptional situation would be in the case of recurrent hydatidiform moles, which raises the possibility of familial biparental hydatidiform mole. This is a pure maternal-effect recessive inherited disorder attributable to mutations of NLRP7 or C6orf221, resulting in a multi-locus epigenetic defect with failure to establish maternal identity at imprinted loci and abnormal expression of imprinted genes.53, 54, 55, 56, 57, 58, 59, 60, 61 Patients with this disorder can have multiple/recurrent complete hydatidiform moles that are morphologically, immunophenotypically, and clinically similar to conventional complete hydatidiform moles, that is, they are p57-negative and appear to have a similar risk of persistent gestational trophoblastic disease, but are characterized by biparental diploidy rather than androgenetic diploidy. These cases are reliably diagnosed as complete hydatidiform moles by concordant morphology and p57 results, but their familial/inherited nature is only established by identifying biparental diploidy per genotyping. Thus, genotyping would be useful in any patient with recurrent hydatidiform moles to determine if these represent the familial form. It is important to recognize that such a biparental form exists so that a genotyping result of biparental diploidy is not used to reject a diagnosis of complete hydatidiform mole when morphology and/or p57 results support that diagnosis. Interestingly, in the current study the one complete hydatidiform mole occurring in a patient with a history of multiple previous hydatidiform moles demonstrated androgenetic diploidy per genotyping, arguing against a diagnosis of a familial hydatidiform mole. Only two cases in our study had the combination of a p57-negative result and biparental diploidy; these did not have any morphology features to suggest a complete hydatidiform mole. Another case with a negative p57 result and unsatisfactory genotyping was quite immature and also lacked any diagnostic features of a complete hydatidiform mole. It is possible these non-molar appearing cases had some genetic or epigenetic alteration affecting p57, leading to a lack of p57 expression. The one early abortus with circumferential trophoblastic hyperplasia, equivocal/suboptimal p57 result, and biparental diploidy per genotyping might be an example of a familial biparental early complete hydatidiform mole; however, there was no known history of recurrent hydatidiform moles or known subsequent ancillary genetic testing to confirm that disorder. Thus, only one potential familial biparental hydatidiform mole was encountered in our study.
The findings in our study also confirm that p57 immunohistochemical analysis is quite useful for recognition of androgenetic/biparental mosaic/chimeric conceptions,24 as discussed in detail in our recent study.44 These include uniformly androgenetic/biparental mosaic specimens without molar features (probably early forms of placental mesenchymal dysplasia, which is characterized by androgenetic/biparental mosaicism and lack of trophoblastic hyperplasia),62 androgenetic/biparental mosaic specimens with a molar component (typically complete hydatidiform mole), and twin gestations comprised of complete hydatidiform mole and non-molar specimen components. Recognition of the discordant and divergent staining patterns in these specimens is the key to correct interpretation of these complex specimens and is necessary for specific microdissection of the different components to assure accurate molecular genotyping.44
As discussed in our previous reproducibility studies,14, 15 the use of the algorithmic approach we advocate represents a compromise between traditional morphological assessment, which has limitations, and genotyping of all cases, which is clearly more costly. As demonstrated herein, the p57 component of the algorithm captures essentially all complete hydatidiform moles, the most important group to readily identify for clinical management purposes. This analysis can be performed in most immunohistochemistry laboratories without the need for highly specialized equipment and expertise, such as that required for genotyping. Genotyping of all p57-positive potentially molar specimens per the algorithm provides for definitive diagnosis of virtually all partial hydatidiform moles and non-molar specimens, allowing for refined management of these entities. However, in the setting of limited resources, use of ancillary techniques can be focused on identifying the entity with the greatest risk for persistent gestational trophoblastic disease, namely complete hydatidiform moles, by selectively applying only p57 immunohistochemistry to assist in diagnosing complete hydatidiform moles and foregoing genotyping for distinction of partial hydatidiform moles from non-molar specimens. For the latter situation, an equivocal diagnosis, such as ‘abnormal villous morphology, partial hydatidiform mole cannot be excluded’, might need to be rendered. As the risk of persistent gestational trophoblastic disease for partial hydatidiform moles is much closer to that of non-molar specimens than complete hydatidiform moles, this may well be acceptable for routine practice, with the understanding that an equivocal diagnosis will potentially lead to clinical management as a partial hydatidiform mole at least for some abbreviated time frame, and that this approach does have accompanying costs (clinic visit, multiple serum beta-HCG levels, contraception), which might well rival the cost of genotyping. It is also worth noting that an apparently unequivocal diagnosis of either partial hydatidiform mole or non-molar specimen established on the basis of morphological assessment alone is not guaranteed to be accurate even when rendered by an experienced gynecologic pathologist. Therefore, the most ideal method of correctly classifying all hydatidiform moles and non-molar specimens is a combined approach including correlation of morphological features, p57 immunohistochemistry, and molecular genotyping. In investigational pursuits, all molar specimens should be evaluated with ancillary techniques to assure rigorous classification of cases, particularly when designed to ascertain risk of persistent gestational trophoblastic disease associated with the various subtypes of hydatidiform moles.
References
Feltmate CM, Growdon WB, Wolfberg AJ et al. Clinical characteristics of persistent gestational trophoblastic neoplasia after partial hydatidiform molar pregnancy. J Reprod Med 2006;51:902–906.
Hancock BW, Nazir K, Everard JE . Persistent gestational trophoblastic neoplasia after partial hydatidiform mole incidence and outcome. J Reprod Med 2006;51:764–766.
Sebire NJ, Fisher RA, Foskett M et al. Risk of recurrent hydatidiform mole and subsequent pregnancy outcome following complete or partial hydatidiform molar pregnancy. BJOG 2003;110:22–26.
Wielsma S, Kerkmeijer L, Bekkers R et al. Persistent trophoblast disease following partial molar pregnancy. Aust N Z J Obstet Gynaecol 2006;46:119–123.
Cheung AN, Khoo US, Lai CY et al. Metastatic trophoblastic disease after an initial diagnosis of partial hydatidiform mole: genotyping and chromosome in situ hybridization analysis. Cancer 2004;100:1411–1417.
Medeiros F, Callahan MJ, Elvin JA et al. Intraplacental choriocarcinoma arising in a second trimester placenta with partial hydatidiform mole. Int J Gynecol Pathol 2008;27:247–251.
Palmieri C, Fisher RA, Sebire NJ et al. Placental site trophoblastic tumour arising from a partial hydatidiform mole. Lancet 2005;366:688.
Seckl MJ, Fisher RA, Salerno G et al. Choriocarcinoma and partial hydatidiform moles. Lancet 2000;356:36–39.
Conran RM, Hitchcock CL, Popek EJ et al. Diagnostic considerations in molar gestations. Hum Pathol 1993;24:41–48.
Fukunaga M, Katabuchi H, Nagasaka T et al. Interobserver and intraobserver variability in the diagnosis of hydatidiform mole. Am J Surg Pathol 2005;29:942–947.
Howat AJ, Beck S, Fox H et al. Can histopathologists reliably diagnose molar pregnancy? J Clin Pathol 1993;46:599–602.
Javey H, Borazjani G, Behmard S et al. Discrepancies in the histological diagnosis of hydatidiform mole. Br J Obstet Gynaecol 1979;86:480–483.
Messerli ML, Parmley T, Woodruff JD et al. Inter- and intra-pathologist variability in the diagnosis of gestational trophoblastic neoplasia. Obstet Gynecol 1987;69:622–626.
Vang R, Gupta M, Wu LS et al. Diagnostic reproducibility of hydatidiform moles: ancillary techniques (p57 immunohistochemistry and molecular genotyping) improve morphologic diagnosis. Am J Surg Pathol 2012;36:443–453.
Gupta M, Vang R, Yemelyanova AV et al. Diagnostic reproducibility of hydatidiform moles: ancillary techniques (p57 immunohistochemistry and molecular genotyping) improve morphologic diagnosis for both recently trained and experienced gynecologic pathologists. Am J Surg Pathol 2012;36:1747–1760.
Chilosi M, Piazzola E, Lestani M et al. Differential expression of p57kip2, a maternally imprinted cdk inhibitor, in normal human placenta and gestational trophoblastic disease. Lab Invest 1998;78:269–276.
Castrillon DH, Sun D, Weremowicz S et al. Discrimination of complete hydatidiform mole from its mimics by immunohistochemistry of the paternally imprinted gene product p57KIP2. Am J Surg Pathol 2001;25:1225–1230.
Fukunaga M . Immunohistochemical characterization of p57(KIP2) expression in early hydatidiform moles. Hum Pathol 2002;33:1188–1192.
Jun SY, Ro JY, Kim KR . p57kip2 is useful in the classification and differential diagnosis of complete and partial hydatidiform moles. Histopathology 2003;43:17–25.
Crisp H, Burton JL, Stewart R et al. Refining the diagnosis of hydatidiform mole: image ploidy analysis and p57KIP2 immunohistochemistry. Histopathology 2003;43:363–373.
Romaguera RL, Rodriguez MM, Bruce JH et al. Molar gestations and hydropic abortions differentiated by p57 immunostaining. Fetal Pediatr Pathol 2004;23:181–190.
Merchant SH, Amin MB, Viswanatha DS et al. p57KIP2 immunohistochemistry in early molar pregnancies: emphasis on its complementary role in the differential diagnosis of hydropic abortuses. Hum Pathol 2005;36:180–186.
Popiolek DA, Yee H, Mittal K et al. Multiplex short tandem repeat DNA analysis confirms the accuracy of p57(KIP2) immunostaining in the diagnosis of complete hydatidiform mole. Hum Pathol 2006;37:1426–1434.
Hoffner L, Dunn J, Esposito N et al. P57KIP2 immunostaining and molecular cytogenetics: combined approach aids in diagnosis of morphologically challenging cases with molar phenotype and in detecting androgenetic cell lines in mosaic/chimeric conceptions. Hum Pathol 2008;39:63–72.
McConnell TG, Murphy KM, Hafez M et al. Diagnosis and subclassification of hydatidiform moles using p57 immunohistochemistry and molecular genotyping: validation and prospective analysis in routine and consultation practice settings with development of an algorithmic approach. Am J Surg Pathol 2009;33:805–817.
Hoffner L, Parks WT, Swerdlow SH et al. Simultaneous detection of imprinted gene expression (p57(KIP2)) and molecular cytogenetics (FICTION) in the evaluation of molar pregnancies. J Reprod Med 2010;55:219–228.
Sarmadi S, Izadi-Mood N, Abbasi A et al. p57KIP2 immunohistochemical expression: a useful diagnostic tool in discrimination between complete hydatidiform mole and its mimics. Arch Gynecol Obstet 2011;283:743–748.
Bell KA, Van d V, Addya K et al. Molecular genetic testing from paraffin-embedded tissue distinguishes nonmolar hydropic abortion from hydatidiform mole. Mol Diagn 1999;4:11–19.
Lai CY, Chan KY, Khoo US et al. Analysis of gestational trophoblastic disease by genotyping and chromosome in situ hybridization. Mod Pathol 2004;17:40–48.
Bifulco C, Johnson C, Hao L et al. Genotypic analysis of hydatidiform mole: an accurate and practical method of diagnosis. Am J Surg Pathol 2008;32:445–451.
Murphy KM, McConnell TG, Hafez MJ et al. Molecular genotyping of hydatidiform moles: analytic validation of a multiplex short tandem repeat assay. J Mol Diagn 2009;11:598–605.
Kajii T, Ohama K . Androgenetic origin of hydatidiform mole. Nature 1977;268:633–634.
Jacobs PA, Wilson CM, Sprenkle JA et al. Mechanism of origin of complete hydatidiform moles. Nature 1980;286:714–716.
Lawler SD, Povey S, Fisher RA et al. Genetic studies on hydatidiform moles. II. The origin of complete moles. Ann Hum Genet 1982;46:209–222.
Ohama K, Kajii T, Okamoto E et al. Dispermic origin of XY hydatidiform moles. Nature 1981;292:551–552.
Lawler SD, Fisher RA, Pickthall VJ et al. Genetic studies on hydatidiform moles. I. The origin of partial moles. Cancer Genet Cytogenet 1982;5:309–320.
Jacobs PA, Szulman AE, Funkhouser J et al. Human triploidy: relationship between parental origin of the additional haploid complement and development of partial hydatidiform mole. Ann Hum Genet 1982;46:223–231.
Zaragoza MV, Surti U, Redline RW et al. Parental origin and phenotype of triploidy in spontaneous abortions: predominance of diandry and association with the partial hydatidiform mole. Am J Hum Genet 2000;66:1807–1820.
Redline RW, Hassold T, Zaragoza BS . Prevalence of the partial molar phenotype in triploidy of maternal and paternal origin. Hum Pathol 1998;29:505–511.
Fallahian M, Sebire NJ, Savage PM et al. Mutations in NLRP7 and KHDC3L confer a complete hydatidiform mole phenotype on digynic triploid conceptions. Hum Mutat 2013;34:301–308.
Chew SH, Perlman EJ, Williams R et al. Morphology and DNA content analysis in the evaluation of first trimester placentas for partial hydatidiform mole (partial hydatidiform mole). Hum Pathol 2000;31:914–924.
Norris-Kirby A, Hagenkord JM, Kshirsagar M et al. Abnormal villous morphology associated with triple trisomy of paternal origin. J Mol Diagn 2010;12:525–529.
Ronnett BM, DeScipio C, Murphy KM . Hydatidiform moles: ancillary techniques to refine diagnosis. Int J Gynecol Pathol 2011;30:101–116.
Lewis GH, DeScipio C, Murphy KM et al. Characterization of androgenetic/biparental mosaic/chimeric conceptions, including those with a molar component: morphology, p57 immunohistochemistry, molecular genotyping, and risk of persistent gestational trophoblastic disease. Int J Gynecol Pathol 2013;32:199–214.
Parazzini F, La VC, Pampallona S . Parental age and risk of complete and partial hydatidiform mole. Br J Obstet Gynaecol 1986;93:582–585.
Sebire NJ, Foskett M, Fisher RA et al. Risk of partial and complete hydatidiform molar pregnancy in relation to maternal age. BJOG 2002;109:99–102.
Rabczynski J, Kochman A, Prudlak E et al. Hydatidiform mole in 59-years-old woman: a case report. Ginekol Pol 2000;71:152–154.
de la FA, Cassignol A, Benkiran L et al. [Invasive hydatiform mole in a postmenopausal woman]. Ann Pathol 2003;23:443–446.
McConnell TG, Norris-Kirby A, Hagenkord JM et al. Complete hydatidiform mole with retained maternal chromosomes 6 and 11. Am J Surg Pathol 2009;33:1409–1415.
DeScipio C, Haley L, Beierl K et al. Diandric triploid hydatidiform mole with loss of maternal chromosome 11. Am J Surg Pathol 2011;35:1586–1591.
Buza N, Hui P . Partial hydatidiform mole: histologic parameters in correlation with DNA genotyping. Int J Gynecol Pathol 2013;32:307–315.
Murphy KM, DeScipio C, Wagenfuehr J et al. Tetraploid partial hydatidiform mole: a case report and review of the literature. Int J Gynecol Pathol 2012;31:73–79.
Helwani MN, Seoud M, Zahed L et al. A familial case of recurrent hydatidiform molar pregnancies with biparental genomic contribution. Hum Genet 1999;105:112–115.
Fisher RA, Khatoon R, Paradinas FJ et al. Repetitive complete hydatidiform mole can be biparental in origin and either male or female. Hum Reprod 2000;15:594–598.
Fisher RA, Hodges MD, Rees HC et al. The maternally transcribed gene p57(KIP2) (CDNK1C) is abnormally expressed in both androgenetic and biparental complete hydatidiform moles. Hum Mol Genet 2002;11:3267–3272.
El-Maarri O, Seoud M, Coullin P et al. Maternal alleles acquiring paternal methylation patterns in biparental complete hydatidiform moles. Hum Mol Genet 2003;12:1405–1413.
Murdoch S, Djuric U, Mazhar B et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 2006;38:300–302.
Van den Veyver I, Al-Hussaini TK . Biparental hydatidiform moles: a maternal effect mutation affecting imprinting in the offspring. Hum Reprod Update 2006;12:233–242.
Kou YC, Shao L, Peng HH et al. A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod 2008;14:33–40.
Hayward BE, De VM, Talati N et al. Genetic and epigenetic analysis of recurrent hydatidiform mole. Hum Mutat 2009;30:E629–E639.
Parry DA, Logan CV, Hayward BE et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet 2011;89:451–458.
Kaiser-Rogers KA, McFadden DE, Livasy CA et al. Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. J Med Genet 2006;43:187–192.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Banet, N., DeScipio, C., Murphy, K. et al. Characteristics of hydatidiform moles: analysis of a prospective series with p57 immunohistochemistry and molecular genotyping. Mod Pathol 27, 238–254 (2014). https://doi.org/10.1038/modpathol.2013.143
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.143
Keywords
This article is cited by
-
Pathogenic role of Twist-1 protein in hydatidiform molar pregnancies and investigation of its potential diagnostic utility in complete moles
Diagnostic Pathology (2023)
-
Gestational trophoblastic disease: an update
Abdominal Radiology (2023)
-
Androgenetic/biparental mosaicism in a diploid mole-like conceptus: report of a case with triple paternal contribution
Virchows Archiv (2023)
-
Recurrent Androgenetic Complete Hydatidiform Moles with p57KIP2-Positive in a Chinese Family
Reproductive Sciences (2022)
-
Refined diagnosis of hydatidiform moles with p57 immunohistochemistry and molecular genotyping: updated analysis of a prospective series of 2217 cases
Modern Pathology (2021)