Abstract
The V600E mutation of BRAF has emerged as both an effective biomarker and therapeutic target for select benign and malignant cutaneous and non-cutaneous human tumors and is typically determined using DNA-based techniques that include allele-specific PCR and direct DNA sequencing. Recently however, the development of new antibodies directed against the V600E protein has opened the door for an easier and more efficient strategy for identifying this mutation. Our present aim was to determine the efficacy of one such antibody, anti-B-Raf (V600E), a mouse monoclonal antibody in which the immunogen is a synthetic peptide derived from the internal region of BRAFV600E. A total of 35 cases of primary cutaneous melanoma were evaluated using a combination of DNA-based techniques that included allele-specific PCR and/or direct DNA sequencing and immunohistochemistry. Cases of papillary thyroid carcinomas (n=5) and colorectal carcinomas (n=5), known to harbor the BRAFV600E mutation, served as positive controls for the study. DNA analyses revealed that 6 of 35 (17%) cases of the primary cutaneous malignant melanoma possessed the BRAFV600E mutation. For immunohistochemical analyses, cytoplasmic positivity with anti-B-Raf was noted in 7 of 35 (20%) cases of primary melanoma and in all 10 positive controls. Statistical analyses of the data demonstrated that the sensitivity of the immunohistochemistry was 100% and specificity was 97%. Findings from the current study support the potential use of immunohistochemistry as an ancillary screening tool to assess the BRAFV600E mutation status in primary cutaneous melanoma.
Similar content being viewed by others
Main
Activating BRAF mutations are found at a high frequency in benign and malignant, cutaneous and non-cutaneous human tumors. Over 90% of these mutations are characterized by a glutamic acid to valine substitution at position 600 (V600E) resulting in a substantial increase in protein kinase activity.1, 2, 3 The BRAFV600E mutation is seen at particularly high rates in primary and metastatic melanoma (20–70%), papillary thyroid carcinoma (40–70%), colorectal carcinoma (5–10%), and in select benign tumors such as melanocytic nevi.2, 4, 5, 6, 7, 8, 9, 10
Given its prevalence and role in increasing tumor cell proliferation and metastases, BRAFV600E has emerged as an important biomarker for clinicians. In early melanomas, the BRAF mutation status has not been shown to affect overall survival; however, in metastatic melanoma, it has been associated with a poorer survival rate.11, 12 With regard to papillary thyroid carcinomas, evidence suggests that molecular testing for BRAFV600E in thyroid fine-needle aspirations can be a useful tool to separate this malignancy from other thyroid nodules.13 The mutation status also serves as a prognostic marker as its presence is associated with higher rates of tumor recurrence and mortality.14 In the case of colorectal carcinomas, BRAFV600E mutation positive patients have significantly shorter progression-free survival when compared with patients with tumors with BRAF wild-type and its presence is shown to impair the therapeutic effect of drugs such as cetuximab and panitumumab.15 The recognition of BRAFV600E mutation and its prognostic value has led to accelerated attempts to utilize it as a target for cancer therapy. Briefly, recent drugs targeted against mutated BRAF (particularly in melanomas) have shown great promise in early clinical trials.10
As it stands today, BRAFV600E mutation status is typically determined by means of DNA-based techniques such as direct DNA Sanger sequencing and allele-specific PCR. However, both of these strategies are expensive and time consuming. Additional confounding issues include the variable quality of DNA extracted from formalin-fixed paraffin-embedded tissue and the obscuring effects of melanin pigment.16 These issues underscore the need for establishing faster and more efficient methods to screen for this mutation. Of particular interest, the development of new antibodies directed against the V600E protein has opened the door for the usage of immunohistochemistry to identify the BRAFV600E mutation. The primary aim of the current study was to determine the efficacy of one such antibody in cases of primary cutaneous melanoma and correlate the results obtained from immunohistochemistry with that evidenced by DNA analyses.
Materials and methods
Sample Selection
This study was approved by Boston University School of Medicine institutional review board (IRB docket #H-31715). Archival tissue with a diagnosis of primary cutaneous malignant melanoma (n=35) were retrieved from the pathology files of the Skin Pathology Laboratory, Boston University School of Medicine, Boston, MA, USA. Inclusion criteria were randomly selected cases of primary cutaneous melanoma from both chronically and intermittently sun-exposed skin with adequate tumoral tissue in the block to allow for genetic and immunohistochemical analyses. Control tissue included cases of thyroid papillary carcinomas (n=5; cases C1–4, 7) and colorectal carcinomas (n=5; cases C5, 6, 8–10) previously determined to harbor the BRAFV600E mutation. Histologic sections of all cases were reviewed by two board-certified dermatopathologists (initial sign-out on all by a dermatopathologist; cases were then re-reviewed, and the diagnoses were confirmed by the senior author). All patient data were de-identified. Demographics of the patients included in the study are detailed in Table 1.
DNA Analyses
DNA was extracted from the sections of formalin-fixed paraffin-embedded primary cutaneous melanoma, papillary thyroid carcinoma and colorectal carcinoma tissues by a simple proteinase K digestion and boiling method without further purification. BRAF mutations were first assayed by allele-specific PCR (codon V600E) by previously described methods.17, 18, 19 Two sets of allele-specific primers (forward and reverse gene coding strand) were designed to duplex with a single pair of GAPDH primers as DNA quality internal control to amplify the mutated allele. Following allele-specific PCR, direct DNA Sanger sequencing was used to confirm the mutation positive cases and a few allele-specific PCR failed cases (as indicated by negative GAPDH PCR). Positive, negative and no-DNA controls were included in each batch of analysis.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue of primary cutaneous malignant melanoma (n=35) and controls were baked at 75 °C for 30 min. Sections were deparaffinized with xylene and rehydrated in a series of decreasing concentrations of ethanol solutions. Heat-induced antigen retrieval was carried out in a DAKO retrieval solution (DAKO, Carpinteria, CA, USA) with a pH of 9.0 in a 98 °C water bath for 12 min. The slides were treated with dual endogenous enzyme block (DAKO) and then incubated with the anti-B-Raf (V600E) mouse monoclonal antibody (NewEast Bioscience, Malvern, PA, USA) at a dilution of 1:100 for 2 h at room temperature. The remaining steps were carried out in a DAKO Autostainer Plus (DAKO). Color development and contrast were achieved using DAB and hematoxylin, respectively.
Cases were marked as positive if they exhibited cytoplasmic staining that was similar in intensity to the positive control in each batch. Any isolated nuclear staining was determined to be negative. All stained slides were reviewed by two observers (first author KF and the senior author MM) in a blinded fashion with respect to genotype.
Results
DNA Analyses
The combined results of allele-specific PCR and direct DNA sequencing revealed that 6 of 35 (17%) of the primary cutaneous melanoma cases (case numbers 9, 10, 11, 20, 22, 33) were positive for the BRAFV600E mutation (Table 2). Of the 29 cases that were negative, 27 of them were shown to have wild-type DNA and the remaining two (cases 2 and 8) possessed mutations other than V600E (case 2 had a glutamic acid to lysine substitution; case 8 had a glutamic acid to methionine substitution). All papillary thyroid carcinoma and colorectal carcinoma cases were positive for BRAFV600E mutation by allele-specific PCR.
Immunohistochemical Analyses
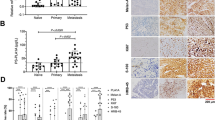
The results of immunohistochemical analyses showed that 7 of 35 (20%) of the primary cutaneous melanoma cases (cases 9, 10, 11, 20, 22, 26, 33) demonstrated positive cytoplasmic staining for the V600E mutated protein (Figure 1; Table 2). In addition to strong cytoplasmic staining, case 20 also possessed nuclear staining. All papillary thyroid carcinoma and colorectal carcinoma cases demonstrated positive cytoplasmic staining for the mutated protein (Figure 1).
Immunohistochemical results for select BRAFV600E mutation positive cases by DNA analyses (case 9=a–c; case 10=d–f; case 20=g–i; papillary thyroid carcinoma control=j–l). (a/d/g/j): Scanning magnification, H&E. (b/e/h/k): High power, H&E. (c/f/i/l): Immunohistochemical stain with the anti-B-Raf (V600E) antibody.
Statistical Analyses
Data analyses revealed that when it came to detecting the BRAFV600E mutational status as determined by direct DNA Sanger sequencing and allele-specific PCR, the anti-B-Raf (V600E) antibody had a sensitivity of 100% and specificity of 97%. All six cases that were positive by DNA analyses were also positive by immunohistochemistry. One case (case 26) was positive only by immunohistochemical analysis.
Discussion
The concept of ‘oncogene addiction’ emphasizes that cancer cells, in contrast to normal cells, rely more heavily on hyperactivated pathways and, therefore, on the oncogenes that drive those pathways for the maintenance of the malignant phenotype.20 A high frequency of BRAF mutations has been previously demonstrated in rapidly changing melanocytic lesions and an increase in frequency of these mutations has also been observed in primary cutaneous malignant melanoma progressing from the radial to the vertical growth phase.21, 22 In light of this, it is not surprising that BRAF has emerged as the ‘Achilles’ heel’ and is a validated therapeutic target in melanoma as BRAFV600E substantially increases protein kinase activity resulting in constitutive BRAF/MEK/ERK signaling.20 To date, select RAF kinase inhibitors are in different phases of several clinical trials, making screening for this mutation of clinical relevance.10
The gold standard for the detection of BRAFV600E is direct DNA sequencing.3 This method takes several days, is expensive and has a relatively low sensitivity in that tumoral population must constitute at least 20% of the tissue sample.23 An additional confounding issue is the suboptimal quality of DNA extracted from formalin-fixed paraffin-embedded tissue, a feature which can necessitate additional rounds of extraction further adding to the delay in turn-round-time. Furthermore, the known interference of melanin in PCR-based techniques as a consequence of its interaction with DNA polymerase can compromise the sensitivity of the procedure.16 Allele-specific PCR circumvents several of the problems associated with direct DNA sequencing in that it has been shown to have the ability to detect roughly 5% of the mutant BRAF in a pool of wild-type, making it particularly attractive in skin samples.24 However, it still does not address the issues of time, cost-effectiveness and DNA extraction.
The advent of monoclonal antibodies on formalin-fixed paraffin-embedded tissue has greatly facilitated the use of immunohistochemistry as a histological adjunct.25 This technique, which is easy to perform and interpret, is also considered to be highly sensitive and specific because the antibodies used are typically directed against the antigen of interest. In other entities associated with genetic abnormalities such as Muir-Torre syndrome, we have previously presented data to support the use of immunohistochemistry as the initial screening test as it has been effective in identifying the loss of one or more mismatch repair proteins in sporadic sebaceous neoplasms.26 An additional minor, yet practically relevant factor relates to the ease of the methodology, which translates to a rapid turn-round-time. Concerning the detection of BRAFV600E, three recent publications highlight the utility of immunohistochemistry using a mutation-specific monoclonal antibody. In the first, Capper et al3 evaluated a total of 47 intracerebral melanoma metastases and 21 primary papillary thyroid carcinomas by techniques that included direct DNA sequencing and immunohistochemistry, albeit with a different antibody (clone VE1), and found a complete correlation between both methodologies. In the second study, using the same antibody Capper et al extended their previous sample size to a larger, more comprehensive cohort of patients with brain metastases and found a correlation between immunohistochemistry and BRAFV600E sequencing in 68/70 cases (∼97%).27 In the third, the same group found a correlation between expression of the protein and the BRAFV600E mutation in 32 cases of hairy cell leukemia.28 However, in each of the latter two studies the authors noted that two cases were positive by immunohistochemistry but negative by sequencing of exon 15 of BRAF and suggested a more sensitive technique to clarify the genetic status. Using a different antibody from that of Capper et al ie anti-B-Raf (V600E), in which the immunogen is a synthetic peptide derived from the internal region of BRAFV600E, we noted that the sensitivity of immunohistochemistry was 100% and specificity was 97%. Of note, only one (case 26) was positive by immunohistochemistry but negative by DNA analyses. As expected, cytoplasmic staining was the pattern observed in all cases. In only one, case 20, additional non-specific nuclear staining was demonstrated. However, given the uniform cytoplasmic staining also observed in this case, it was scored as a positive.
The focus of the current study was not on evaluating the frequency of BRAFV600E mutations in primary cutaneous melanoma, but on ascertaining the utility of a mutation-specific monoclonal antibody. Therefore, our cases were not selected specifically to verify mutation rates. However, it should be noted that our reported incidence of BRAFV600E in primary cutaneous melanoma is relatively low, albeit not unprecedented. Studies on the prevalence of BRAF mutations in melanoma indicate that they are most commonly seen in melanomas from skin sites subject to sun exposure, with a relatively higher prevalence being noted in those that are intermittently sun-exposed vs continually sun-exposed.7 This might account for the slightly lower frequency of BRAFV600E in the current cohort as a proportion of our cases were from continually sun-exposed sites (cases 1, 2, 7, 10–12, 15, 21, 23, 26, 30). Furthermore, it has been previously shown using the same technique that the frequency of BRAFV600E mutations in primary cutaneous melanoma can vary greatly with incidences as low as 30% being reported.11 Our own previous experience has shown that the frequency could be even lower.8
In spite of the low incidence rate, findings from this preliminary study shed light on the potential use of immunohistochemistry with the anti-B-Raf (V600E) antibody as an ancillary screening tool to assess the BRAFV600E mutation status in primary cutaneous melanoma. Further, in support of the specificity of this antibody, two cases in our cohort possessed mutations other than V600E (case 2 had a glutamic acid to lysine substitution while case 8 had a glutamic acid to methionine substitution) and were negative by immunohistochemistry (Figure 2). Ours is not the first study to document V600 mutations other than V600E. In one previous study, the same glutamic acid to lysine substitution was noted and found in a higher proportion (7%) of cases.29 While the precise significance of these other mutations is unclear, it further validates the specificity of the antibody for the BRAFV600E mutation.
Cases with BRAF mutation other than V600E (case 2=a–d; case 8=e–h). (a) Direct DNA sequencing demonstrating the glutamic acid to lysine substitution (GTG to AAG). (b/f) Scanning magnification, H&E. (c/g) High power, H&E. (d/h) Immunohistochemical stain with the anti-B-Raf (V600E) antibody (brown staining seen is melanin). (e) Direct DNA sequencing demonstrating the glutamic acid to methionine substitution (GTG to ATG).
While we observed the staining intensity to be somewhat variable, a consistent feature of all seven positive cases was that the staining was confined to lesional melanocytes. In contrast to DNA-based techniques, this allows for the distinction to be made between populations harboring the mutant allele vs those harboring wild-type. From a scientific perspective, this feature facilitates insights into the biology of nevic lesions particularly those in which the melanoma arises in the setting of a banal or congenital nevus. Additional studies on a larger cohort are required to validate our findings and for immunohistochemistry to supplement or replace DNA-based techniques in identifying BRAFV600E in primary cutaneous melanoma. Given the similar results seen in both the thyroid papillary carcinoma and colorectal carcinoma controls, we are encouraged about the potential of this mutation-specific antibody.
References
Wan P, Garnett M, Roe SM et al Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116:855–867.
Wajapeyee N, Serra R, Zhu X et al Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 2008;132:363–374.
Capper D, Preusser M, Habel A et al Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol 2011;122:11–19.
Yuen S, Davies H, Chan T et al Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 2002;62:6451–6455.
Cohen Y, Xing M, Mambo E et al BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst 2003;95:625–627.
Kimura E, Nikiforova M, Zhu Z et al High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res 2003;63:1454–1457.
DeCarlo K, Yang S, Emley A et al Oncogenic BRAF-positive dysplastic nevi and the tumor suppressor IGFBP7—challenging the concept of dysplastic nevi as precursor lesions? Hum Pathol 2010;41:886–894.
Dadzie OE, Yang S, Emley A et al RAS and RAF mutations in banal melanocytic aggregates contiguous with primary cutaneous melanoma: clues to melanomagenesis. Br J Dermatol 2008;1–8.
Miller D, Emley A, Yang S et al Mixed versus pure variants of desmoplastic melanoma: a genetic and immunohistochemical appraisal. Mod Pathol 2011;1–11.
Arkenau HT, Kefford R, Long GV . Targeting BRAF for patients with melanoma. Br J Cancer 2011;104:392–398.
Shinozaki M, Fujimoto A, Morton D et al Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res 2004;10:1753–1757.
Long G, Menzies A, Nagrial A et al Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol 2011;29:1239–1246.
Nikiforova YE, Steward DL, Robinson-Smith TM et al Molecular testing for mutations in improving the fine-needle aspiration diagnosis of thyroid nodules. J Clin Endocrinol Metab 2009;94:2092–2098.
Nikiforova YE . Molecular diagnostics of thyroid tumors. Arch Pathol Lab Med 2011;135:569–577.
Di Nicolantonio F, Martini M, Molinari F et al Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705–5712.
Eckhart L, Bach J, Ban J et al Melanin binds reversibly to thermostable DNA polymerase and inhibits its activity. Biochem Biophys Res Commun 2000;271:726–730.
Pollock PM, Harper UL, Hansen KS et al High frequency of BRAF mutations in nevi. Nat Genet 2003;33:19–20.
Yang S, Farraye FA, Mack C et al BRAF and KRAS mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol 2004;28:1452–1459.
O’Brien M, Yang S, Mack C et al Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and tradtional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 2006;30:1491–1501.
Weinstein IB, Joe A . Oncogene addiction. Cancer Res 2008;68:3077–3080.
Kumar R, Angelini S, Snellman E et al BRAF mutations are common somatic events in melanocytic nevi. J Invest Dermatol 2004;122:342–348.
Greene VR, Johnson MM, Grimm EA et al Frequencies of NRAS and BRAF mutations increase from the radial to the vertical growth phase in cutaneous melanoma. J Invest Dermatol 2009;129:1483–1488.
Lee HJ, Choi J, Hwang T et al Detection of BRAF mutations in thyroid nodules by allele-specific PCR using a dual priming oligonucleotide system. Am J Clin Pathol 2010;133:802–808.
Panka D, Sullivan R, Mier J . An inexpensive specific and highly sensitive protocol to detect the BRAFV600E mutation in melanoma tumor biopsies and blood. Melanoma Res 2010;20:401–407.
Abbas O, Mahalingam M . Cutaneous sebaceous neoplasms as markers of Muir-Torre syndrome: a diagnostic algorithm. J Cutan Pathol 2009;36:613–619.
Chhibber V, Dresser K, Mahalingam M . MSH-6: extending the reliability of immunohistochemistry as a screening tool in Muir-Torre syndrome. Mod Pathol 2008;21:159–164.
Capper D, Berghoff AS, Magerle M et al Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol 2012;123:223–233.
Andrulis M, Penzel R, Weichart W et al Application of a BRAF V600E mutation-specific antibody for the diagnosis of hairy cell leukemia. Am J Surg Pathol 2012;36:1–5.
Smalley KSM, Xiao M, Villanueva J et al CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene 2009;28:85–94.
Acknowledgements
We would like to acknowledge Neweast Biosciences for providing the antibody used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Feller, J., Yang, S. & Mahalingam, M. Immunohistochemistry with a mutation-specific monoclonal antibody as a screening tool for the BRAFV600E mutational status in primary cutaneous malignant melanoma. Mod Pathol 26, 414–420 (2013). https://doi.org/10.1038/modpathol.2012.168
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.168
Keywords
This article is cited by
-
Is immunohistochemistry of BRAF V600E useful as a screening tool and during progression disease of melanoma patients?
BMC Cancer (2016)
-
NRAS Q61R , BRAF V600E immunohistochemistry: a concomitant tool for mutation screening in melanomas
Diagnostic Pathology (2015)
-
Immunohistochemistry is highly sensitive and specific for the detection of NRASQ61R mutation in melanoma
Modern Pathology (2015)
-
Basics of Immunohistochemistry
Journal of Investigative Dermatology (2015)
-
Mycoplasma gallisepticum MGA_0676 is a membrane-associated cytotoxic nuclease with a staphylococcal nuclease region essential for nuclear translocation and apoptosis induction in chicken cells
Applied Microbiology and Biotechnology (2015)