Abstract
Mucoepidermoid carcinoma is the most common salivary gland malignancy, and includes a spectrum of lesions ranging from non-aggressive low-grade tumors to aggressive high-grade tumors. To further characterize this heterogeneous group of tumors we have performed a comprehensive analysis of copy number alterations and CRTC1–MAML2 fusion status in a series of 28 mucoepidermoid carcinomas. The CRTC1–MAML2 fusion was detected by RT-PCR or fluorescence in situ hybridization in 18 of 28 mucoepidermoid carcinomas (64%). All 15 low-grade tumors were fusion-positive whereas only 3 of 13 high-grade tumors were fusion-positive. High-resolution array-based comparative genomic hybridization revealed that fusion-positive tumors had significantly fewer copy number alterations/tumor compared with fusion-negative tumors (1.5 vs 9.5; P=0.002). Twelve of 18 fusion-positive tumors had normal genomic profiles whereas only 1 out of 10 fusion-negative tumors lacked copy number alterations. The profiles of fusion-positive and fusion-negative tumors were very similar to those of low- and high-grade tumors. Thus, low-grade mucoepidermoid carcinomas had significantly fewer copy number alterations/tumor compared with high-grade mucoepidermoid carcinomas (0.7 vs 8.6; P<0.0001). The most frequent copy number alterations detected were losses of 18q12.2-qter (including the tumor suppressor genes DCC, SMAD4, and GALR1), 9p21.3 (including the tumor suppressor genes CDKN2A/B), 6q22.1-q23.1, and 8pter-p12.1, and gains of 8q24.3 (including the oncogene MAFA), 11q12.3-q13.2, 3q26.1-q28, 19p13.2-p13.11, and 8q11.1-q12.2 (including the oncogenes LYN, MOS, and PLAG1). On the basis of these results we propose that mucoepidermoid carcinoma may be subdivided in (i) low-grade, fusion-positive mucoepidermoid carcinomas with no or few genomic imbalances and favorable prognosis, (ii) high-grade, fusion-positive mucoepidermoid carcinomas with multiple genomic imbalances and unfavorable prognosis, and (iii) a heterogeneous group of high-grade, fusion-negative adenocarcinomas with multiple genomic imbalances and unfavorable outcome. Taken together, our studies indicate that molecular genetic analysis can be a useful adjunct to histologic scoring of mucoepidermoid carcinoma and may lead to development of new clinical guidelines for management of these patients.
Similar content being viewed by others
Main
Mucoepidermoid carcinoma is the most common type of salivary gland carcinoma and may be found in both the major and minor salivary glands.1, 2, 3 The histological classification of AFIP adopted by the WHO,3, 4, 5 grade mucoepidermoid carcinoma based on histopathological features including a cystic component, nerve invasion, necrosis, mitotic activity, and cytological pleomorphism. It is recognized that high-grade mucoepidermoid carcinomas are associated with a high risk of recurrences, metastases, and tumor-related deaths, whereas low-grade mucoepidermoid carcinomas usually have an excellent prognosis and only rarely metastasize.4, 5, 6, 7 Nevertheless, all tumors with histologic appearances defined as mucoepidermoid carcinoma are considered malignant.8 Although efforts have been made to identify clinically useful biomarkers for grading and prognostication9, 10, 11, 12 there is yet no unifying concept on how to classify mucoepidermoid carcinomas.
We previously identified a recurrent t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma.13, 14 Subsequent studies revealed that it results in a CRTC1–MAML2 fusion in which the N-terminal Notch-binding domain of the coactivator MAML2 (Mastermind-like 2) is replaced by the CREB-binding domain of CRTC1.15, 16 An important molecular consequence of the fusion is the activation of cAMP/CREB target genes (Enlund et al., unpublished data).17, 18 Several studies have now confirmed the initial report by Behboudi et al19 demonstrating that the CRTC1–MAML2 fusion primarily occurs in low-grade mucoepidermoid carcinomas with a favorable clinical outcome.12, 13, 20, 21 A second gene fusion involving the sarcoma-associated EWSR1 gene and the stem cell regulator POU5F1 was recently identified in mucoepidermoid carcinomas with a more immature morphology compared with MAML2 positive tumors.22
Except for the CRTC1–MAML2 fusion, little is known about other genomic rearrangements of importance for the genesis and progression of mucoepidermoid carcinoma.23, 24 To identify such alterations, we performed a genome-wide, array-based comparative genomic hybridization (arrayCGH) study of a series of 28 mucoepidermoid carcinomas. Our results demonstrate that low- and high-grade mucoepidermoid carcinomas have different genomic profiles and CRTC1–MAML2 fusion status. Low-grade tumors have few or no genomic imbalances and are fusion-positive whereas high-grade tumors have numerous genomic imbalances and are often fusion-negative. The results provide additional evidence supporting that mucoepidermoid carcinoma is a heterogeneous tumor entity and that genetic biomarkers may be useful in identifying subgroups with different clinical outcomes.
Materials and methods
Patients and Tissue Specimens
A total of 28 mucoepidermoid carcinomas were analyzed in this study, including 13 archival formalin-fixed paraffin-embedded tumors and 15 fresh-frozen tumors. The samples were retrieved from the files of the Department of Pathology, Haartman Institute, University of Helsinki, Finland, the Department of Pathology, Sahlgrenska University Hospital, Gothenburg, Sweden, and from the Department of Oral Pathology, Guy’s Hospital, London, UK. The diagnoses were reviewed by three pathologists and histopathological grading of the tumors was performed according to the WHO classification.3 Other differential diagnosis of high-grade mucoepidermoid carcinomas, such as salivary duct carcinomas and adenosquamous carcinomas were excluded to the best of the authors' ability using morphology and immunohistochemical criteria described in the WHO classification.3 Clinical follow-up data were obtained from the patients medical records. Ethical approval was obtained from the National Supervisory Authority for Welfare and Health in Finland (VALVIRA) (Dnro 1451/32/300/04 and 425/05.01.00.06/2009), and ethical approvals from the Ethics Committee of Helsinki University Hospital, Finland (Dnro 410/E9/05), Guy’s Research Ethics Committee in London, UK (reference 02/10/14),and from the regional ethics committee in Gothenburg, Sweden (D-no: 178-08).
arrayCGH Analysis
Tumor samples were trimmed to remove adjacent non-neoplastic tissues. Genomic DNA was extracted from formalin-fixed paraffin-embedded and fresh-frozen tumor tissues as previously described.25, 26 A pool of normal female or male genomic DNAs obtained from peripheral blood cells (each from five normal individuals) was used as reference DNA. arrayCGH analysis was performed using the Human Genome CGH Microarray 44K and 244K oligonucleotide arrays (Agilent Technologies, Palo Alto, CA) as previously described and as recommended by the manufacturer.26 The slides were subsequently scanned using the Agilent microarray confocal scanner G2565AA (Agilent Technologies). Images were analyzed using the Feature Extraction software (v7.5; Agilent Technologies) with intensity-dependent linear normalization to reduce inter-experimental variation.
Data analysis was carried out using Nexus Copy Number software v.4.1 (BioDiscovery, El Segundo, CA). Nexus Copy Number uses the Rank segmentation algorithm to define non-random regions of copy number alterations across the genome. Sex chromosomes were excluded from the analysis. The significance threshold for segmentation was set to P=1.0E−7 and the log2 ratio thresholds for gain and loss were 0.4 and −0.3, respectively. The log2 ratio thresholds for high copy number gain/amplification and homozygous deletion were 1.0 and −1.0, respectively. A copy number alteration was considered recurrent if three or more tumor samples carried the same copy number alteration with a p-value of less than 0.05. Each aberration was checked manually to confirm the accuracy of the call. Regions partially or completely covered by a previously reported copy number variation (Database of Genomic Variants; http://dgvbeta.tcag.ca/dgv/app/news?ref=NCBI36/hg18) were excluded from the analysis.27
RT-PCR Analysis
The CRTC1–MAML2 fusion transcript was detected by nested RT-PCR using primers located in exon 1 of CRTC1 and exon 2 of MAML2 as previously described.16, 19
Fluorescence In Situ Hybridization (FISH) Analysis
FISH analyses for detection of the CRTC1–MAML2 and EWSR1–POU5F1 gene fusions were performed on 3 μm formalin-fixed paraffin-embedded sections using dual-color break-apart rearrangement probes for the MAML2 (ZytoVision GmbH, Bremerhaven, Germany) and EWSR1 genes (Vysis, Downer’s Grove, IL) as previously described.22, 28
To validate recurrent copy number alterations detected by arrayCGH we performed FISH analyses using locus-specific probes for MALT1 located at 18q21.32 (MALT1 Break-apart probe; Cytocell, Cambridge, UK) and MECOM located at 3q26.2 (MECOM/RUNX1 t(3;21) Fusion-probe; Kreatech Diagnostics, Amsterdam, The Netherlands). Locus-specific probes for CCND1 (IGH/ CCND1 Translocation Probe; Cytocell) and HER2 (Vysis) were used to confirm amplifications involving 11q and 17q sequences. The protocols for pretreatment, hybridization, and posthybridization washes were essentially as recommended by the manufacturers. Fluorescence signals were digitized, processed, and analyzed using the CytoVision image analysis system (Applied Imaging International, Newcastle-Upon-Tyne, UK). Thirty to 300 nuclei were scored from each case.
Results
Clinical and Histopathological Characteristics
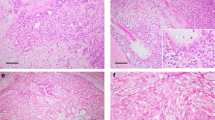
The clinical and histopathological data for all mucoepidermoid carcinoma patients are detailed in Table 1. Fifteen of the tumors were classified as low-grade and 13 as high-grade mucoepidermoid carcinomas (Figure 1). The mean age of patients with low-grade tumors was 47 years (range 7–73 years) whereas the corresponding figure for those with high-grade tumors was 64 years (range 29–85 years). Nine of the 13 patients with high-grade mucoepidermoid carcinomas developed distant metastases whereas none of the patients with low-grade tumors developed metastasis during the follow-up period (range 6–23 years). Recurrences were found in 5 of the 28 patients. They included both fusion-positive and negative tumors as well as low- and high-grade tumors.
Photomicrographs of mucoepidermoid carcinomas. (a) Low-power view of case 6 showing a low-grade CRTC1–MAML2 fusion-positive mucoepidermoid carcinoma. (b) High-power view of case 18 showing a high-grade fusion-positive mucoepidermoid carcinoma. (c) High-power view of case 23 showing a high-grade fusion-negative mucoepidermoid carcinoma.
CRTC1–MAML2 and EWSR1–POU5F1 Fusion Gene Status
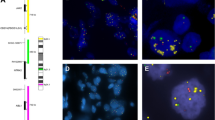
The CRTC1–MAML2 fusion oncogene was detected by RT-PCR or FISH in 18 of the 28 mucoepidermoid carcinoma cases (64%). All 15 low-grade tumors were fusion-positive (Figure 2a) and 10 of 13 high-grade tumors (77%) were fusion-negative (Table 1 and Figure 2b). The CRTC1–MAML2-negative mucoepidermoid carcinomas were also analyzed by FISH for the EWSR1–POU5F1 fusion using an EWSR1 dual-color break-apart probe. None of the 10 MAML2-negative tumors showed a rearrangement consistent with an EWSR1 gene fusion (data not shown).
FISH analyses of CRTC1–MAML2 (a, b) and copy number alterations detected by arrayCGH (c–f) in mucoepidermoid carcinoma. FISH analysis of the CRTC1–MAML2 fusion gene using a MAML2 break-apart probe in a fusion-positive (a) (case 3; separated green and red signals) and fusion-negative (b) (case 21; fused green and red signals) mucoepidermoid carcinoma. arrayCGH and FISH analyses showing loss of the MALT1 locus at 18q21 (one red signal/nucleus) in case 18 (c), gain of the MECOM locus at 3q26 (multiple red signals/nucleus) in case 19 (d), amplification of CCND1 at 11q13 (multiple single and clustered red signals) in case 19 (e), and amplification of ERBB2 at 17q12 (multiple single and clustered red signals) in case 27 (f). The locations of the genes on the respective chromosomes are indicated by an asterics. Losses are indicated by red lines to the left of each chromosome and gains with blue lines to the right of each chromosome.
Genomic Profiles in Mucoepidermoid Carcinoma
A detailed description of all copy number alterations identified are presented in Table 1. A total of 122 copy number alterations were recorded in 15 tumors (Figure 3a). The remaining 13 tumors had no detectable copy number alterations. The number of genomic imbalances per tumor was significantly lower in fusion-positive tumors compared with fusion-negative tumors (1.5 vs 9.5; P=0.002) (Figure 3b). Twelve of the 18 fusion-positive tumors had normal genomic profiles whereas only one out of 10 fusion-negative tumors lacked copy number alterations. Two high-grade, fusion-positive mucoepidermoid carcinomas had copy number alterations with breakpoints in 11q21 and 19p13 consistent with CRTC1–MAML2 fusions generated by t(11;19) translocations. The genomic profiles of fusion-positive and fusion-negative tumors were very similar to those of low- and high-grade tumors. Thus, low-grade mucoepidermoid carcinomas had significantly fewer copy number alterations per tumor compared with high-grade mucoepidermoid carcinomas (0.7 vs 8.6; P<0.0001) (Figure 3c). The mean number of imbalances for high-grade fusion-positive tumors (n=3) was 5.7 compared with 9.5 in high-grade fusion-negative tumors (n=10). Twelve of 15 low-grade mucoepidermoid carcinomas had apparently normal genomic profiles. The three remaining tumors showed a total of 10 copy number alterations. In contrast, 12 of the 13 high-grade mucoepidermoid carcinomas contained 1–29 copy number alterations per tumor (Table 1).
Genome-wide frequency plot of copy number alterations in 28 mucoepidermoid carcinomas. A total of 122 copy number alterations were detected across the genome (sex chromosomes excluded). Losses (red) were more common than gains (blue) (a). Distribution of the different copy number alterations in CRTC1–MAML2-positive (Fus +) and -negative (Fus −) tumors (b) and in low-grade (LG) and high-grade (HG) mucoepidermoid carcinomas. The number of genomic imbalances per tumor was significantly lower in fusion-positive tumors compared with fusion-negative tumors (1.5 vs 9.5; P=0.002). Similarly, LG mucoepidermoid carcinomas had significantly fewer copy number alterations per tumor compared with HG mucoepidermoid carcinomas (0.7 vs 8.6; P<0.0001). Note that the profiles of fusion-positive and LG tumors are very similar and that the profiles of fusion-negative and HG tumors are almost identical.
At least 13 recurrent minimal common regions of copy number losses and gains were identified in the 28 mucoepidermoid carcinomas (Table 2). A list of the genes located in these minimal common regions is shown in Supporting Information Table S1. The most frequently lost regions were 18q12.2-qter (eight cases; including the tumor suppressor genes DCC, SMAD4, and GALR1), 9p21.3 (seven cases; including the tumor suppressor genes CDKN2A/B), 6q22.1-q23.1 (four cases), 8pter-p12.1 (four cases), 5q13.2-q15 (three cases), and 4p (three cases) (Figures 4a and b). The deletions involving 9p21.3 were found in both high- (n=6) and low-grade (n=1) mucoepidermoid carcinomas and included an approximately 60 kb minimal common region containing CDKN2A/B. Two tumors had homozygous deletions of 2.3 and 6.2 Mb, respectively, involving the CDKN2A/B genes. The most frequently gained regions were 8q24.3 (seven cases; including the MAFA candidate oncogene), 11q12.3-q13.2 (six cases), 3q26.1-q28 (five cases), 19p13.2-p13.11 (five cases), 8q11.1-q12.2 (four cases; including the LYN, MOS, and PLAG1 oncogenes), 5pter-p15.31 (three cases), and 9q33.3-q34.3 (three cases) (Figure 4c). Recurrent gain of one chromosome 19 was seen in three tumors, two of which were fusion-positive. Three high-grade, fusion-negative mucoepidermoid carcinomas had amplifications with single continuous amplicons at 7q11.23-q21.2, 11q13.2-q13.5, and 17q12-q21.2, respectively (Table 1). A list of all genes located within these three amplicons is shown in Supporting Information Table S2. The 11q13.2-13.5 amplicon included the CCND1 gene and the 17q12-q21.2 the ERBB2 gene.
We also analyzed separately the arrayCGH profiles for the 18 CRTC1–MAML2 fusion-positive mucoepidermoid carcinoma cases. Six tumors contained 1–8 copy number alterations per tumor whereas 12 cases had normal genomic profiles. The following copy number alterations were detected in ≥3 mucoepidermoid carcinoma samples: gains of 11q12.2-q13.2 (four cases), 19p13.2-p13.11 (four cases), 8q24.3 (three cases), and 9q33.3-q34.3 (three cases).
Validation of Copy Number Alterations Using FISH
To confirm the copy number alterations detected by arrayCGH, we performed FISH analysis of tumors with losses involving 18q, gains of 3q, and amplifications of 11q13.2-13.5 and 17q12-q21.2. Using a probe for MALT1, located at 18q21.32, we could confirm loss of this locus in all six tumors analyzed (cases 11, 18, 20, 21, 23, and 25). Sixty to seventy-five percent of the tumor cells showed a single MALT1 signal consistent with loss of one 18q allele (Figure 2c). Similarly, the FISH analysis confirmed gain of the MECOM gene located at 3q26.2 in all three cases analyzed (cases 11, 19, and 25). Fifty to ninety-five percent of the tumor cell nuclei displayed three to seven signals consistent with gains of this gene complex (Figure 2d). Amplifications of CCND1 and ERBB2 were also confirmed by FISH in cases 19 and 27, respectively. More than 90% of the tumor cell nuclei in both cases showed multiple signals and/or clusters of signals consistent with amplification of CCND1 (Figure 2e) and ERBB2 (Figure 2f).
Discussion
Using high-resolution arrayCGH, FISH, and RT-PCR we have performed a comprehensive analysis of genomic imbalances and gene fusion status in a series of histologically and clinically well-characterized mucoepidermoid carcinomas. To our knowledge, this is the largest and most comprehensive array-based CGH study performed on mucoepidermoid carcinoma. Previous analyses of genomic imbalances in mucoepidermoid carcinoma using CGH are limited to one chromosomal-based CGH-study of 16 tumors23 and one array-based study of 15 tumors24 (Supporting Information Table S3). Both studies showed similar percentages of CRTC1–MAML2 fusion-positive tumors (58% vs 61%). The most prominent finding in these studies was loss of the CDKN2A gene in a subset of fusion-positive tumors and that loss of this gene was associated with an unfavorable prognosis.24
In the present study comprising 28 tumors, 12 of 15 low-grade mucoepidermoid carcinomas had apparently normal genomic profiles, which is consistent with the well-known non-aggressive clinical behavior of most low-grade mucoepidermoid carcinomas.6, 10 None of these tumors metastasized and only two recurred during the follow-up period. Moreover, all low-grade tumors were positive for the CRTC1–MAML2 gene fusion, whereas only three of the 13 high-grade mucoepidermoid carcinomas were fusion-positive.7, 12, 19, 20, 21 The remaining 10 fusion-negative, high-grade tumors were also negative for the EWSR1–POU5F1 fusion previously identified in high-grade mucoepidermoid carcinomas.22 These findings support the notion that low-grade mucoepidermoid carcinomas are fusion-positive and genetically stable tumors with few genomic imbalances. The fact that CRTC1–MAML2 is a potent oncogene with effects on critical signaling pathways (Enlund et al, unpublished data)17, 29 might at least partly explain why these tumors contain relatively few copy number alterations.
To identify genetic events that may cooperate with CRTC1–MAML2 in mucoepidermoid carcinoma tumorigenesis or disease progression we performed an unbiased search for recurrent copy number alterations in fusion-positive mucoepidermoid carcinomas. In the six fusion-positive tumors with genomic imbalances we detected four copy number alterations that were found in ≥3 cases, that is gains of 11q12.2-q13.2, 19p13.2-p13.11, 8q24.3, and 9q33.3-q34.3. Two of these gains, 11q12.2-q13.2 and 19p13.2-p13.11, were only recurrent in fusion-positive tumors, suggesting that they may harbor genes which can cooperate with CRTC1–MAML2 in mucoepidermoid carcinoma tumorigenesis. Of interest, it was recently shown that gain of 11q13.1 in fusion-positive mucoepidermoid carcinomas is associated with unfavorable prognosis.23 The 11q12.2-q13.2 and 19p13.2-p13.11 segments contain several cancer-associated genes such as RIN1, FOSL1, and VEGFB (11q12.2-q13.2) and JUNB and JUND (19p13.2-p13.11) that are duplicated/rearranged and/or overexpressed in various forms of epithelial cancers.30, 31, 32 Whether the 11q and 19p gains in mucoepidermoid carcinoma target any of these genes remains, however, to be shown.
The most frequent copy number alterations detected in the 28 mucoepidermoid carcinomas were losses of 18q12.2-qter, 9p21.3, 6q22.1-q23.1, and 8pter-p12.1, and gains of 8q24.3, 11q12.3-q13.2, 3q26.1-q28, 19p13.2-p13.11, and 8q11.1-q12.2. The frequencies of these copy number alterations varied from 11 to 29%, suggesting that there are no high-frequency copy number alterations in mucoepidermoid carcinoma and that a given copy number alteration therefore is likely to be of pathogenetic importance only for a subset of patients. The most common copy number alteration was loss of 18q12.2-qter found in 29% of the mucoepidermoid carcinomas. Heterozygous loss of 18q has been reported in several types of carcinomas, for example colon cancer,33 pancreatic carcinoma,34 and head and neck squamous cell carcinoma.35 These losses, including the tumor suppressor genes SMAD4, DCC, and GALR1, are associated with tumor progression and unfavorable prognosis.33, 34, 36 Of interest, all but one of our mucoepidermoid carcinomas with loss of 18q were high-grade tumors that developed metastases (five cases) and/or recurrences (three cases).
The second most common copy number loss included an approximately 60 kb minimal common region within 9p21.3, harboring the tumor suppressor genes CDKN2A/B. In two of the tumors the deletions were homozygous. The 9p deletions were detected in one low- and six high-grade mucoepidermoid carcinomas. All five high-grade tumors with known follow-up data developed metastases and/or recurrences. These findings are partly in agreement with recent data showing that fusion-positive mucoepidermoid carcinomas with inactivating CDKN2A deletions have an unfavorable prognosis.24 Taken together, the present and previous studies show that CDKN2A deletions do occur in both fusion-positive and fusion-negative mucoepidermoid carcinomas and that they are associated with an unfavorable outcome. Deletion or hypermethylation of CDKN2A is a frequent oncogenic event in various types of carcinomas, including for example lung cancer, head and neck squamous cell carcinoma, and salivary duct carcinoma.37, 38, 39 Further studies will be needed to confirm the significance of CDKN2A deletions in mucoepidermoid carcinomas with and without CRTC1–MAML2 gene fusion.
The most frequent copy number gain was a 1.4 Mb minimal common region in 8q24.3 that was gained in seven tumors. This region is also frequently gained in several other types of carcinomas.40, 41 An interesting candidate target gene of these gains is MAFA, an oncogene with transforming properties that is overexpressed in multiple myeloma and a subtype of T-cell lymphoma.42 We also detected recurrent gains of 8q11.1-q12.2 (harboring the LYN, MOS, and PLAG1 oncogenes) in four high-grade tumors. This is of special interest because we recently showed that gain of a 1.4 Mb segment in 8q12.1, containing the PLAG1 gene, is of importance for malignant transformation of benign salivary pleomorphic adenomas.43, 44
In summary, we have shown that low-grade mucoepidermoid carcinomas have normal or near-normal genomic profiles, express the CRTC1–MAML2 fusion, and have a favorable clinical outcome. In contrast, the majority of high-grade tumors had multiple genomic imbalances, were negative for the CRTC1–MAML2 fusion, and developed frequent metastasis and/or recurrences. On the basis of these results we propose a subdivision of mucoepidermoid carcinomas, in (i) low-grade, fusion-positive mucoepidermoid carcinomas with no or few genomic imbalances and a favorable prognosis, (ii) high-grade, fusion-positive mucoepidermoid carcinomas with multiple genomic imbalances and an unfavorable prognosis, and (iii) high-grade, fusion-negative tumors with multiple genomic imbalances and an unfavorable clinical outcome. The latter tumors likely constitute a heterogeneous group of diverse high-grade adenocarcinomas with some mucoepidermoid carcinoma-like morphologic features. Taken together, the present and previous studies indicate that molecular genetic analysis can be a useful adjunct to histologic scoring of mucoepidermoid carcinoma and may lead to development of new clinical guidelines for management of these patients and ultimately also to new therapeutic strategies.
References
Goode RK, Auclair PL, Ellis GL . Mucoepidermoid carcinoma of the major salivary glands: clinical and histopathologic analysis of 234 cases with evaluation of grading criteria. Cancer 1998;82:1217–1224.
Guzzo M, Andreola S, Sirizzotti G et al. Mucoepidermoid carcinoma of the salivary glands: clinicopathologic review of 108 patients treated at the National Cancer Institute of Milan. Ann Surg Oncol 2002;9:688–695.
Goode RK, El-Naggar AK . Mucoepidermoid carcinoma In: Barnes L, Eveson JW, Reichart P, Sidransky D, (eds) World Health Organization Classification of Tumours: Pathology and Genetics of Head and Neck Tumours. IARC Press: Lyon, France, 2005, pp 219–220.
Ellis GL, Auclair PL . AFIP Atlas of Tumor Pathology, Series 4, Fascicle 9. Tumors of the Salivary Glands. American Registry of Pathology: Washington, DC, 2008, pp 173–196.
Auclair PL, Goode RK, Ellis GL . Mucoepidermoid carcinoma of intraoral salivary gland. Evaluation and application of grading criteria in 143 cases. Cancer 1992;69:2021–2030.
Aro K, Leivo I, Mäkitie AA . Management and outcome of patients with mucoepidermoid carcinoma of major salivary gland origin: a single institution's 30-year experience. Laryngoscope 2008;118:258–262.
Seethala RR, Dacic S, Cieply K et al. A reappraisal of the MECT1/MAML2 translocation in salivary mucoepidermoid carcinomas. Am J Surg Pathol 2010;34:1106–1121.
Seifert G . Mucoepidermoid carcinoma in a salivary duct cyst of the parotid gland. Contribution to the development of tumours in salivary gland cysts. Pathol Res Pract 1996;192:1211–1217.
Skalova A, Lehtonen H, von Boguslawsky K et al. Prognostic significance of cell proliferation in mucoepidermoid carcinomas of the salivary gland: clinicopathological study using MIB 1 antibody in paraffin sections. Hum Pathol 1994;25:929–935.
Brandwein MS, Ivanov K, Wallace DI et al. Mucoepidermoid carcinoma: a clinicopathologic study of 80 patients with special reference to histological grading. Am J Surg Pathol 2001;25:835–845.
Nance MA, Seethala RR, Wang Y et al. Treatment and survival outcomes based on histologic grading in patients with head and neck mucoepidermoid carcinoma. Cancer 2008;113:2082–2089.
Miyabe S, Okabe M, Nagatsuka H et al. Prognostic significance of p27Kip1, Ki-67, and CRTC1-MAML2 fusion transcript in mucoepidermoid carcinoma: a molecular and clinicopathologic study of 101 cases. J Oral Maxillofac Surg 2009;67:1432–1441.
Nordkvist A, Gustafsson H, Juberg-Ode M et al. Recurrent rearrangements of 11q14-22 in mucoepidermoid carcinoma. Cancer Genet Cytogenet 1994;74:77–83.
Mitelman F, Johansson B, Mertens F, (eds.). Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancerhttp://cgap.nci.nih.gov/Chromosomes/Mitelman 2012.
Tonon G, Modi S, Wu L et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet 2003;33:208–213.
Enlund F, Behboudi A, Andrén Y et al. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin's tumors. Exp Cell Res 2004;292:21–28.
Coxon A, Rozenblum E, Park YS et al. Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res 2005;65:7137–7144.
Wu L, Liu J, Gao P et al. Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J 2005;24:2391–2402.
Behboudi A, Enlund F, Winnes M et al. Molecular classification of mucoepidermoid carcinomas—prognostic significance of the MECT1–MAML2 fusion oncogene. Genes Chromosomes Cancer 2006;45:470–481.
Okabe M, Miyabe S, Nagatsuka H et al. MECT1-MAML2 fusion transcript defines a favorable subset of mucoepidermoid carcinoma. Clin Cancer Res 2006;12:3902–3907.
Tirado Y, Williams MD, Hanna EY et al. CRTC1/MAML2 fusion transcript in high-grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin’s tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer 2007;46:708–715.
Möller E, Stenman G, Mandahl N et al. POU5F1, encoding a key regulator of stem cell pluripotency, is fused to EWSR1 in hidradenoma of the skin and mucoepidermoid carcinoma of the salivary glands. J Pathol 2008;215:78–86.
Verdorfer I, Fehr A, Bullerdiek J et al. Chromosomal imbalances, 11q21 rearrangement and MECT1-MAML2 fusion transcript in mucoepidermoid carcinomas of the salivary gland. Oncol Rep 2009;22:305–311.
Anzick SL, Chen WD, Park Y et al. Unfavorable prognosis of CRTC1-MAML2 positive mucoepidermoid tumors with CDKN2A deletions. Genes Chromosomes Cancer 2010;39:59–69.
Järvinen AK, Autio R, Kilpinen S et al. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer 2008;47:500–509.
Persson F, Winnes M, Andrén Y et al. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene 2008;27:3072–3080.
Iafrate AJ, Feuk L, Rivera MN et al. Detection of large-scale variation in the human genome. Nat Genet 2004;36:949–951.
Winnes M, Mölne L, Suurküla M et al. Frequent fusion of the CRTC1 and MAML2 genes in clear cell variants of cutaneous hidradenomas. Genes Chromosomes Cancer 2007;46:559–563.
Komiya T, Park Y, Modi S et al. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene 2006;25:6128–6132.
Fischer C, Mazzone M, Jonckx B et al. FLT1 and its ligands VEGFB and PlGF: drug targets for anti-angiogenic therapy? Nat Rev Cancer 2008;8:942–956.
Tomshine JC, Severson SR, Wigle DA et al. Cell proliferation and epidermal growth factor signaling in non-small cell lung adenocarcinoma cell lines are dependent on Rin1. J Biol Chem 2009;284:26331–26339.
Lopez-Bergami P, Lau E, Ronai Z . Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer 2010;10:65–76.
Jen J, Kim H, Piantadosi S et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med 1994;331:213–221.
Hahn SA, Schutte M, Hoque AT et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996;271:350–353.
Takebayashi S, Hickson A, Ogawa T et al. Loss of chromosome arm 18q with tumor progression in head and neck squamous cancer. Genes Chromosomes Cancer 2004;41:145–154.
Misawa K, Ueda Y, Kanazawa T et al. Epigenetic inactivation of galanin receptor 1 in head and neck cancer. Clin Cancer Res 2008;14:7604–7613.
Rocco JW, Sidransky D . p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res 2001;264:42–55.
Baylin SB, Ohm JE . Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006;6:107–116.
Brock MV, Hooker CM, Ota-Machida E et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008;358:1118–1128.
van Duin M, van Marion R, Vissers KJ et al. High-resolution array comparative genomic hybridization of chromosome 8q: evaluation of putative progression markers for gastroesophageal junction adenocarcinomas. Cytogenet Genome Res 2007;118:130–137.
Parris TZ, Danielsson A, Nemes S et al. Clinical implications of gene dosage and gene expression patterns in diploid breast carcinoma. Clin Cancer Res 2010;16:3860–3874.
Eychène A, Rocques N, Pouponnot C . A new MAFia in cancer. Nat Rev Cancer 2008;8:683–693.
Persson F, Andrén Y, Winnes M et al. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer 2009;48:69–82.
Stenman G, Andersson MK, Andrén Y . New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell Cycle 2010;9:2986–2995.
Acknowledgements
This study was supported by grants from the Finnish Cancer Society, Helsinki University Hospital Fund, Maritza and Reino Salonen Foundation, Finska Läkaresällskapet (Helsinki), Swedish Cancer Society, BioCARE—a National Strategic Research Program at University of Gothenburg, and the Royal Academy of Arts and Sciences in Gothenburg.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Jee, K., Persson, M., Heikinheimo, K. et al. Genomic profiles and CRTC1–MAML2 fusion distinguish different subtypes of mucoepidermoid carcinoma. Mod Pathol 26, 213–222 (2013). https://doi.org/10.1038/modpathol.2012.154
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.154
Keywords
This article is cited by
-
Mucoepidermoid carcinoma (MEC) and adenosquamous carcinoma (ASC), the same or different entities?
Modern Pathology (2022)
-
Development of head and neck pathology in Europe
Virchows Archiv (2022)
-
Whole-exome sequencing reveals the etiology of the rare primary hepatic mucoepidermoid carcinoma
Diagnostic Pathology (2021)
-
Mucoepidermoid carcinoma of the salivary glands revisited with special reference to histologic grading and CRTC1/3-MAML2 genotyping
Virchows Archiv (2021)
-
MAML2 rearrangement as a useful diagnostic marker discriminating between Warthin tumour and Warthin-like mucoepidermoid carcinoma
Virchows Archiv (2020)