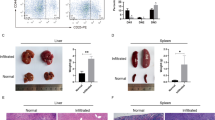
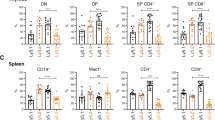
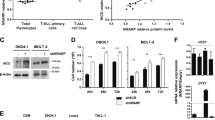
Abstract
RUNX3, runt-domain transcription factor, is a master regulator of gene expression in major developmental pathways. It acts as a tumor suppressor in many cancers but is oncogenic in certain tumors. We observed upregulation of RUNX3 mRNA and protein expression in nasal-type extranodal natural killer (NK)/T-cell lymphoma (NKTL) patient samples and NKTL cell lines compared to normal NK cells. RUNX3 silenced NKTL cells showed increased apoptosis and reduced cell proliferation. Potential binding sites for MYC were identified in the RUNX3 enhancer region. Chromatin immunoprecipitation–quantitative PCR revealed binding activity between MYC and RUNX3. Co-transfection of the MYC expression vector with RUNX3 enhancer reporter plasmid resulted in activation of RUNX3 enhancer indicating that MYC positively regulates RUNX3 transcription in NKTL cell lines. Treatment with a small-molecule MYC inhibitor (JQ1) caused significant downregulation of MYC and RUNX3, leading to apoptosis in NKTL cells. The growth inhibition resulting from depletion of MYC by JQ1 was rescued by ectopic MYC expression. In summary, our study identified RUNX3 overexpression in NKTL with functional oncogenic properties. We further delineate that MYC may be an important upstream driver of RUNX3 upregulation and since MYC is upregulated in NKTL, further study on the employment of MYC inhibition as a therapeutic strategy is warranted.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ito Y . Oncogenic potential of the RUNX gene family: 'Overview'. Oncogene 2004; 23: 4198–4208.
Ito Y . RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res 2008; 99: 33–76.
Kundu M, Compton S, Garrett-Beal L, Stacy T, Starost MF, Eckhaus M et al. Runx1 deficiency predisposes mice to T-lymphoblastic lymphoma. Blood 2005; 106: 3621–3624.
Leong DT, Lim J, Goh X, Pratap J, Pereira BP, Kwok HS et al. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res 2010; 12: R89.
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res 2005; 65: 7743–7750.
Lee CW, Ito K, Ito Y . Role of RUNX3 in bone morphogenetic protein signaling in colorectal cancer. Cancer Res 2010; 70: 4243–4252.
Fujii S, Ito K, Ito Y, Ochiai A . Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem 2008; 283: 17324–17332.
Cameron ER, Neil JC . The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene 2004; 23: 4308–4314.
Salto-Tellez M, Peh BK, Ito K, Tan SH, Chong PY, Han HC et al. RUNX3 protein is overexpressed in human basal cell carcinomas. Oncogene 2006; 25: 7646–7649.
Tsunematsu T, Kudo Y, Iizuka S, Ogawa I, Fujita T, Kurihara H et al. RUNX3 has an oncogenic role in head and neck cancer. PLoS ONE 2009; 4: e5892.
Lee CWL, Chuang LSH, Kimura S, Lai SK, Ong CW, Yan B et al. RUNX3 functions as an oncogene in ovarian cancer. Gynecol Oncol 2011; 122: 410–417.
Bledsoe KL, McGee-Lawrence ME, Camilleri ET, Wang X, Riester SM, van Wijnen AJ et al. RUNX3 facilitates growth of Ewing sarcoma cells. J Cell Physiol 2014; 229: 2049–2056.
Chan JKC, Quintanilla-Martinez L, Ferry JA, Peh SC . Extranodal NK/T-cell lymphoma, nasal type. In: Swerdlow SH et al WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th edn. IRAC Press: Lyon, France, 2008, pp 285–288.
Ohno S, Sato T, Kohu K, Takeda K, Okumura K, Satake M et al. Runx proteins are involved in regulation of CD122, Ly49 family and IFN-gamma expression during NK cell differentiation. Int Immunol 2008; 20: 71–79.
Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, Groner Y . Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLoS ONE 2013; 8: e80467.
Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 2009; 206: 51–59.
Ng SB, Selvarajan V, Huang G, Zhou J, Feldman AL, Law M et al. Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J Pathol 2011; 223: 496–510.
Ng SB, Yan J, Huang G, Selvarajan V, Tay JL, Lin B et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood 2011; 118: 4919–4929.
Ito Y, Bae SC, Chuang LS . The RUNX family: developmental regulators in cancer. Nat Rev Cancer 2015; 15: 81–95.
Puig-Kröger A, Aguilera-Montilla N, Martínez-Nuñez R, Domínguez-Soto A, Sánchez-Cabo F, Martín-Gayo E et al. The novel RUNX3/p33 isoform is induced upon monocyte-derived dendritic cell maturation and downregulates IL-8 expression. Immunobiology 2010; 215: 812–820.
Chi XZ, Kim J, Lee YH, Lee JW, Lee KS, Wee H et al. Runt-related transcription factor RUNX3 is a target of MDM2-mediated ubiquitination. Cancer Res 2009; 69: 8111–8119.
Lee YS . Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer cell 2013; 24: 603–616.
Yamada C, Ozaki T, Ando K, Suenaga Y, Inoue K-i, Ito Y et al. RUNX3 modulates DNA damage-mediated phosphorylation of tumor suppressor p53 at Ser-15 and acts as a co-activator for p53. J Biol Chem 2010; 285: 16693–16703.
Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood 2010; 115: 1226–1237.
Iqbal J, Kucuk C, Deleeuw RJ, Srivastava G, Tam W, Geng H et al. Genomic analyses reveal global functional alterations that promote tumor growth and novel tumor suppressor genes in natural killer-cell malignancies. Leukemia 2009; 23: 1139–1151.
Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet 2015; 47: 1061–1066.
Koo GC, Tan SY, Tang T, Poon SL, Allen GE, Tan L et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov 2012; 2: 591–597.
Stewart M, MacKay N, Cameron ER, Neil JC . The common retroviral insertion locus Dsi1 maps 30 kilobases upstream of the P1 promoter of the murine Runx3/Cbfa3/Aml2 gene. J Virol 2002; 76: 4364–4369.
Stewart M . Proviral insertions induce the expression of bone-specific isoforms of PEBP2[alpha]A (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci USA 1997; 94: 8646–8651.
Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005; 21: 2933–2942.
Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O et al. Selective inhibition of BET bromodomains. Nature 2010; 468: 1067–1073.
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–917.
Gregory MA, Hann SR . c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol 2000; 20: 2423–2435.
Kudo Y, Tsunematsu T, Takata T . Oncogenic role of RUNX3 in head and neck cancer. J Cell Biochem 2011; 112: 387–393.
Nevadunsky NS, Barbieri JS, Kwong J, Merritt MA, Welch WR, Berkowitz RS et al. RUNX3 protein is overexpressed in human epithelial ovarian cancer. Gynecol Oncol 2009; 112: 325–330.
Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS, Kang SG et al. Expression of RUNX3 in skin cancers. Clin Exp Dermatol 2011; 36: 769–774.
Lacayo NJ, Meshinchi S, Kinnunen P, Yu R, Wang Y, Stuber CM et al. Gene expression profiles at diagnosis in de novo childhood AML patients identify FLT3 mutations with good clinical outcomes. Blood 2004; 104: 2646–2654.
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res 2006; 66: 6512–6520.
Lee YS, Lee JW, Jang JW, Chi XZ, Kim JH, Li YH et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell 2013; 24: 603–616.
Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, Jenkins A et al. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res 2006; 66: 2195–2201.
Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron ER et al. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene 2001; 20: 295–302.
Boyd RL, Hugo P . Towards an integrated view of thymopoiesis. Immunol Today 1991; 12: 71–79.
Sugimoto KJ, Kawamata N, Sakajiri S, Oshimi K . Molecular analysis of oncogenes, ras family genes (N-ras, K-ras, H-ras), myc family genes (c-myc, N-myc) and mdm2 in natural killer cell neoplasms. Jpn J Cancer Res 2002; 93: 1270–1277.
Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B . The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol 1999; 73: 4481–4484.
Dirmeier U, Hoffmann R, Kilger E, Schultheiss U, Briseno C, Gires O et al. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 2005; 24: 1711–1717.
Ng SB, Khoury JD . Epstein-Barr virus in lymphoproliferative processes: an update for the diagnostic pathologist. Adv Anat Pathol 2009; 16: 40–55.
Coppo P, Gouilleux-Gruart V, Huang Y, Bouhlal H, Bouamar H, Bouchet S et al. STAT3 transcription factor is constitutively activated and is oncogenic in nasal-type NK/T-cell lymphoma. Leukemia 2009; 23: 1667–1678.
Yu H, Kortylewski M, Pardoll D . Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 2007; 7: 41–51.
Yan J, Ng SB, Tay JL, Lin B, Koh TL, Tan J et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 2013; 121: 4512–4520.
Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151: 68–79.
Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014; 511: 488–492.
Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 2014; 511: 483–487.
Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA 2011; 108: 16669–16674.
Acknowledgements
S-BN was supported by the Singapore Ministry of Health’s National Medical Research Council Transition Award (NMRC/TA/0020/2013). This work is supported in part by Singapore Ministry of Education Academic Research Fund Tier 1 (WBS No: R-179-000-046-112). W-JC was supported by the Singapore Ministry of Health’s National Medical Research Council Clinician Scientist Investigator Award. Ethics approval was obtained from IRB, National University of Singapore, ID: 10‐107.
Author contributions
W-JC conceived and designed study, and analyzed the data; S-BN conceived and designed the study, analyzed the data and wrote the paper. VS performed experiments and wrote the paper; GSSN, MO, JY, DC-CV and YI provided vital reagents and interpreted findings; T-HC performed bioinformatics analysis; MFH, MS-T, S-NC and SF constructed TMA, performed IHC and DIF; NS maintained and contributed cell lines.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Selvarajan, V., Osato, M., Nah, G. et al. RUNX3 is oncogenic in natural killer/T-cell lymphoma and is transcriptionally regulated by MYC. Leukemia 31, 2219–2227 (2017). https://doi.org/10.1038/leu.2017.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.40
This article is cited by
-
RUNX transcription factors: biological functions and implications in cancer
Clinical and Experimental Medicine (2024)
-
Super-enhancer-driven TOX2 mediates oncogenesis in Natural Killer/T Cell Lymphoma
Molecular Cancer (2023)
-
Clinical significance of determining the hypermethylation of the RUNX3 gene promoter and its cohypermethylation with the BRCA1 gene for patients with breast cancer
Journal of Cancer Research and Clinical Oncology (2023)
-
EZH2 regulates a SETDB1/ΔNp63α axis via RUNX3 to drive a cancer stem cell phenotype in squamous cell carcinoma
Oncogene (2022)
-
Effect of circular RNA, mmu_circ_0000296, on neuronal apoptosis in chronic cerebral ischaemia via the miR-194-5p/Runx3/Sirt1 axis
Cell Death Discovery (2021)