Abstract
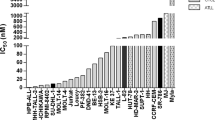
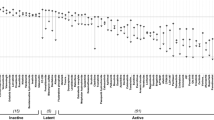
T-cell prolymphocytic leukemia (T-PLL) is a rare and aggressive neoplasm of mature T-cells with an urgent need for rationally designed therapies to address its notoriously chemo-refractory behavior. The median survival of T-PLL patients is <2 years and clinical trials are difficult to execute. Here we systematically explored the diversity of drug responses in T-PLL patient samples using an ex vivo drug sensitivity and resistance testing platform and correlated the findings with somatic mutations and gene expression profiles. Intriguingly, all T-PLL samples were sensitive to the cyclin-dependent kinase inhibitor SNS-032, which overcame stromal-cell-mediated protection and elicited robust p53-activation and apoptosis. Across all patients, the most effective classes of compounds were histone deacetylase, phosphoinositide-3 kinase/AKT/mammalian target of rapamycin, heat-shock protein 90 and BH3-family protein inhibitors as well as p53 activators, indicating previously unexplored, novel targeted approaches for treating T-PLL. Although Janus-activated kinase–signal transducer and activator of transcription factor (JAK-STAT) pathway mutations were common in T-PLL (71% of patients), JAK-STAT inhibitor responses were not directly linked to those or other T-PLL-specific lesions. Overall, we found that genetic markers do not readily translate into novel effective therapeutic vulnerabilities. In conclusion, novel classes of compounds with high efficacy in T-PLL were discovered with the comprehensive ex vivo drug screening platform warranting further studies of synergisms and clinical testing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Herling M, Khoury JD, Washington LT, Duvic M, Keating MJ, Jones D . A systematic approach to diagnosis of mature T-cell leukemias reveals heterogeneity among WHO categories. Blood 2004; 104: 328–335.
Herling M, Patel KA, Teitell MA, Konopleva M, Ravandi F, Kobayashi R et al. High TCL1 expression and intact T-cell receptor signaling define a hyperproliferative subset of T-cell prolymphocytic leukemia. Blood 2008; 111: 328–337.
Dearden C . How I treat prolymphocytic leukemia. Blood 2012; 120: 538–551.
Hopfinger G, Busch R, Pflug N, Weit N, Westermann A, Fink AM et al. Sequential chemoimmunotherapy of fludarabine, mitoxantrone, and cyclophosphamide induction followed by alemtuzumab consolidation is effective in T-cell prolymphocytic leukemia. Cancer 2013; 119: 2258–2267.
Krishnan B, Else M, Tjonnfjord GE, Cazin B, Carney D, Carter J et al. Stem cell transplantation after alemtuzumab in T-cell prolymphocytic leukaemia results in longer survival than after alemtuzumab alone: a multicentre retrospective study. Br J Haematol 2010; 149: 907–910.
Herling M . Are we improving the outcome for patients with T-cell prolymphocytic leukemia by allogeneic stem cell transplantation? Eur J Haematol 2015; 94: 191–192.
Wiktor-Jedrzejczak W, Dearden C, de Wreede L, van Biezen A, Brinch L, Leblond V et al. Hematopoietic stem cell transplantation in T-prolymphocytic leukemia: a retrospective study from the European Group for Blood and Marrow Transplantation and the Royal Marsden Consortium. Leukemia 2012; 26: 972–976.
Stengel A, Kern W, Zenger M, Perglerova K, Schnittger S, Haferlach T et al. Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker. Genes Chromosomes Cancer 2016; 55: 82–94.
Yokohama A, Saitoh A, Nakahashi H, Mitsui T, Koiso H, Kim Y et al. TCL1A gene involvement in T-cell prolymphocytic leukemia in Japanese patients. Int J Hematol 2012; 95: 77–85.
Stern MH, Soulier J, Rosenzwajg M, Nakahara K, Canki-Klain N, Aurias A et al. MTCP-1: a novel gene on the human chromosome Xq28 translocated to the T cell receptor alpha/delta locus in mature T cell proliferations. Oncogene 1993; 8: 2475–2483.
Virgilio L, Lazzeri C, Bichi R, Nibu K, Narducci MG, Russo G et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci USA 1998; 95: 3885–3889.
Gritti C, Dastot H, Soulier J, Janin A, Daniel MT, Madani A et al. Transgenic mice for MTCP1 develop T-cell prolymphocytic leukemia. Blood 1998; 92: 368–373.
Kiel MJ, Velusamy T, Rolland D, Sahasrabuddhe AA, Chung F, Bailey NG et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood 2014; 124: 1460–1472.
Hu Z, Medeiros LJ, Fang L, Sun Y, Tang Z, Tang G et al. Prognostic significance of cytogenetic abnormalities in T-cell prolymphocytic leukemia. Am J Hematol 2017; 92: 441–447.
Delgado P, Starshak P, Rao N, Tirado CA . A comprehensive update on molecular and cytogenetic abnormalities in T-cell prolymphocytic leukemia (T-PLL). J Assoc Genet Technol 2012; 38: 193–198.
Durig J, Bug S, Klein-Hitpass L, Boes T, Jons T, Martin-Subero JI et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32). Leukemia 2007; 21: 2153–2163.
Gaudio E, Spizzo R, Paduano F, Luo Z, Efanov A, Palamarchuk A et al. Tcl1 interacts with Atm and enhances NF-kappaB activation in hematologic malignancies. Blood 2012; 119: 180–187.
Bellanger D, Jacquemin V, Chopin M, Pierron G, Bernard OA, Ghysdael J et al. Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia 2014; 28: 417–419.
Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov 2013; 3: 1416–1429.
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016; 127: 2375–2390.
Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med 2012; 366: 1905–1913.
Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol 2011; 12: R6.
Maljaie SH, Brito-Babapulle V, Matutes E, Hiorns LR, De Schouwer PJ, Catovsky D . Expression of c-myc oncoprotein in chronic T cell leukemias. Leukemia 1995; 9: 1694–1699.
Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ et al. Inconsistency in large pharmacogenomic studies. Nature 2013; 504: 389–393.
Haverty PM, Lin E, Tan J, Yu Y, Lam B, Lianoglou S et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature 2016; 533: 333–337.
Dose M, Emmanuel AO, Chaumeil J, Zhang J, Sun T, Germar K et al. beta-Catenin induces T-cell transformation by promoting genomic instability. Proc Natl Acad Sci USA 2014; 111: 391–396.
Keating KE, Gueven N, Watters D, Rodemann HP, Lavin MF . Transcriptional downregulation of ATM by EGF is defective in ataxia-telangiectasia cells expressing mutant protein. Oncogene 2001; 20: 4281–4290.
Chen R, Wierda WG, Chubb S, Hawtin RE, Fox JA, Keating MJ et al. Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood 2009; 113: 4637–4645.
Li L, Pongtornpipat P, Tiutan T, Kendrick SL, Park S, Persky DO et al. Synergistic induction of apoptosis in high-risk DLBCL by BCL2 inhibition with ABT-199 combined with pharmacologic loss of MCL1. Leukemia 2015; 29: 1702–1712.
Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev 2014; 28: 1800–1814.
Warner K, Weit N, Crispatzu G, Admirand J, Jones D, Herling M . T-cell receptor signaling in peripheral T-cell lymphoma - a review of patterns of alterations in a central growth regulatory pathway. Curr Hematol Malig Rep 2013; 8: 163–172.
Bergmann AK, Schneppenheim S, Seifert M, Betts MJ, Haake A, Lopez C et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer 2014; 53: 309–316.
Lopez C, Bergmann AK, Paul U, Murga Penas EM, Nagel I, Betts MJ et al. Genes encoding members of the JAK-STAT pathway or epigenetic regulators are recurrently mutated in T-cell prolymphocytic leukaemia. Br J Haematol 2016; 173: 265–273.
Huang CY, Lin YC, Hsiao WY, Liao FH, Huang PY, Tan TH . DUSP4 deficiency enhances CD25 expression and CD4+ T-cell proliferation without impeding T-cell development. Eur J Immunol 2012; 42: 476–488.
Hsiao WY, Lin YC, Liao FH, Chan YC, Huang CY . Dual-specificity phosphatase 4 regulates STAT5 protein stability and helper T cell polarization. PLoS One 2015; 10: e0145880.
Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun 2015; 6: 6025.
Herling M, Patel KA, Weit N, Lilienthal N, Hallek M, Keating MJ et al. High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood 2009; 114: 4675–4686.
Heath EI, Bible K, Martell RE, Adelman DC, Lorusso PM . A phase 1 study of SNS-032 (formerly BMS-387032), a potent inhibitor of cyclin-dependent kinases 2, 7 and 9 administered as a single oral dose and weekly infusion in patients with metastatic refractory solid tumors. Invest New Drugs 2008; 26: 59–65.
Le Toriellec E, Despouy G, Pierron G, Gaye N, Joiner M, Bellanger D et al. Haploinsufficiency of CDKN1B contributes to leukemogenesis in T-cell prolymphocytic leukemia. Blood 2008; 111: 2321–2328.
Tong WG, Chen R, Plunkett W, Siegel D, Sinha R, Harvey RD et al. Phase I and pharmacologic study of SNS-032, a potent and selective Cdk2, 7, and 9 inhibitor, in patients with advanced chronic lymphocytic leukemia and multiple myeloma. J Clin Oncol 2010; 28: 3015–3022.
Brito-Babapulle V, Hamoudi R, Matutes E, Watson S, Kaczmarek P, Maljaie H et al. p53 allele deletion and protein accumulation occurs in the absence of p53 gene mutation in T-prolymphocytic leukaemia and Sezary syndrome. Br J Haematol 2000; 110: 180–187.
Hasanali ZS, Saroya BS, Stuart A, Shimko S, Evans J, Vinod Shah M et al. Epigenetic therapy overcomes treatment resistance in T cell prolymphocytic leukemia. Sci Transl Med 2015; 7: 293ra102.
Cayrol F, Praditsuktavorn P, Fernando TM, Kwiatkowski N, Marullo R, Calvo-Vidal MN et al. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat Commun 2017; 8: 14290.
Pemovska T, Johnson E, Kontro M, Repasky GA, Chen J, Wells P et al. Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature 2015; 519: 102–105.
Heinrich T, Rengstl B, Muik A, Petkova M, Schmid F, Wistinghausen R et al. Mature T-cell lymphomagenesis induced by retroviral insertional activation of Janus kinase 1. Mol Ther 2013; 21: 1160–1168.
Spinner S, Crispatzu G, Yi JH, Munkhbaatar E, Mayer P, Hockendorf U et al. Re-activation of mitochondrial apoptosis inhibits T-cell lymphoma survival and treatment resistance. Leukemia 2016; 30: 1520–1530.
Warner K, Crispatzu G, Al-Ghaili N, Weit N, Florou V, You MJ et al. Models for mature T-cell lymphomas—a critical appraisal of experimental systems and their contribution to current T-cell tumorigenic concepts. Crit Rev Oncol Hematol 2013; 88: 680–695.
Kirouac DC, Saez-Rodriguez J, Swantek J, Burke JM, Lauffenburger DA, Sorger PK . Creating and analyzing pathway and protein interaction compendia for modelling signal transduction networks. BMC Syst Biol 2012; 6: 29.
Zaman N, Li L, Jaramillo ML, Sun Z, Tibiche C, Banville M et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell Rep 2013; 5: 216–223.
Acknowledgements
This work was supported by the Academy of Finland, the Finnish Cancer Societies, Finnish Cancer Institute, Instrumentarium Science Foundation, Biomedicum Helsinki Foundation, Sigrid Juselius Foundation, European Regional Development Fund, Signe and Ane Gyllenberg Foundation, Swedish Cultural Foundation, Blood Disease Foundation the Finnish Cultural Foundation. MH and AS were supported by the DFG Research Unit FOR1961 (CONTROL-T; HE3553/4-2), by the Köln Fortune program by the Fritz Thyssen Foundation (10.15.2.034MN) and by the José Carreras Leukemia Foundation (DJCLS 03F/2016). TA and JT were supported by European Union’s Horizon 2020 research and innovation program (grant Agreement No. 634143, MedBioinformatics). MO, WH and TZ were supported by the European Commission’s Horizon 2020 Project SOUND. We thank Professor Kimmo Porkka and Dr Caroline Heckman for their scientific input; Dr Esa Jantunen, Dr Marja Pyörälä, Dr Marjut Kauppila, Dr Maija Itälä-Remes and Dr Veli Kairisto for providing patient information and the personnel at the Hematology Research Unit Helsinki and FIMM for their expert clinical and technical assistance.
Author contributions
EIA and SM designed the study, coordinated the project, analyzed the data and wrote the paper. EIA and SL performed sequence analysis and validated mutations. EIA, SP, OD, TP and PP designed and performed the functional experiments. BY, SAK, LH, SE, GC, MO and JPM designed and performed the bioinformatics analysis. LS, AS, SP, HZ, DB, AL, KT, CC-M, EF, SK, ERS, TS and TB provided patient samples and participated in the laboratory studies. SA, PE, OK, WD, M-HS, WH, KW, JT, TA, TZ and MH participated in the study design, data analysis and contributed to write the paper. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Labcyte, Inc. and FIMM/University of Helsinki have a collaboration agreement on the utilization of Labcyte’s acoustic dispensing technologies. CC-M is an employee of IMMED.S.L. SM has received honoraria and research funding from Novartis, Pfizer and Bristol-Myers Squibb (not related to this study). KW has received honoraria and research funding from Novartis and Pfizer (not related to this study). The other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Andersson, E., Pützer, S., Yadav, B. et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia 32, 774–787 (2018). https://doi.org/10.1038/leu.2017.252
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.252
This article is cited by
-
Prolymphocytic Leukaemia: an Update on Biology and Treatment
Current Oncology Reports (2024)
-
All that glitters is not LGL Leukemia
Leukemia (2022)
-
Identification of novel STAT5B mutations and characterization of TCRβ signatures in CD4+ T-cell large granular lymphocyte leukemia
Blood Cancer Journal (2022)
-
Venetoclax treatment of patients with relapsed T-cell prolymphocytic leukemia
Blood Cancer Journal (2021)
-
Reinstated p53 response and high anti-T-cell leukemia activity by the novel alkylating deacetylase inhibitor tinostamustine
Leukemia (2020)