Abstract
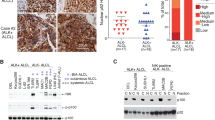
Classical Hodgkin lymphoma (cHL) and anaplastic large cell lymphoma (ALCL) feature high expression of activator protein-1 (AP-1) transcription factors, which regulate various physiological processes but also promote lymphomagenesis. The AP-1 factor basic leucine zipper transcription factor, ATF-like 3 (BATF3), is highly transcribed in cHL and ALCL; however, its functional importance in lymphomagenesis is unknown. Here we show that proto-typical CD30+ lymphomas, namely cHL (21/30) and primary mediastinal B-cell lymphoma (8/9), but also CD30+ diffuse large B-cell lymphoma (15/20) frequently express BATF3 protein. Mass spectrometry and co-immunoprecipitation established interactions of BATF3 with JUN and JUNB in cHL and ALCL lines. BATF3 knockdown using short hairpin RNAs was toxic for cHL and ALCL lines, reducing their proliferation and survival. We identified MYC as a critical BATF3 target and confirmed binding of BATF3 to the MYC promoter. JAK/STAT signaling regulated BATF3 expression, as chemical JAK2 inhibition reduced and interleukin 13 stimulation induced BATF3 expression in cHL lines. Chromatin immunoprecipitation substantiated a direct regulation of BATF3 by STAT proteins in cHL and ALCL lines. In conclusion, we identified STAT-mediated BATF3 expression that is essential for lymphoma cell survival and promoted MYC activity in cHL and ALCL, hence we recognized a new oncogenic axis in these lymphomas.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Schmitz R, Stanelle J, Hansmann M-L, Küppers R . Pathogenesis of classical and lymphocyte-predominant Hodgkin lymphoma. Annu Rev Pathol 2009; 4: 151–174.
Kanzler H, Küppers R, Hansmann ML, Rajewsky K . Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996; 184: 1495–1505.
Küppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci USA 1994; 91: 10962–10966.
Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J 2002; 21: 4104–4113.
Eckerle S, Brune V, Doring C, Tiacci E, Bohle V, Sundstrom C et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia 2009; 23: 2129–2138.
Hapgood G, Savage KJ . The biology and management of systemic anaplastic large cell lymphoma. Blood 2015; 126: 17–25.
Stein H, Mason DY, Gerdes J, O'Connor N, Wainscoat J, Pallesen G et al. The expression of the Hodgkin's disease associated antigen Ki-1 in reactive and neoplastic lymphoid tissue: evidence that Reed-Sternberg cells and histiocytic malignancies are derived from activated lymphoid cells. Blood 1985; 66: 848–858.
Eferl R, Wagner EF . AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003; 3: 859–868.
Drakos E, Leventaki V, Schlette EJ, Jones D, Lin P, Medeiros LJ et al. c-Jun expression and activation are restricted to CD30+ lymphoproliferative disorders. Am J Surg Pathol 2007; 31: 447–453.
Rassidakis GZ, Thomaides A, Atwell C, Ford R, Jones D, Claret FX et al. JunB expression is a common feature of CD30+ lymphomas and lymphomatoid papulosis. Mod Pathol 2005; 18: 1365–1370.
Laimer D, Dolznig H, Kollmann K, Vesely PW, Schlederer M, Merkel O et al. PDGFR blockade is a rational and effective therapy for NPM-ALK-driven lymphomas. Nat Med 2012; 18: 1699–1704.
Rodig SJ, Ouyang J, Juszczynski P, Currie T, Law K, Neuberg DS et al. AP1-dependent galectin-1 expression delineates classical hodgkin and anaplastic large cell lymphomas from other lymphoid malignancies with shared molecular features. Clin Cancer Res 2008; 14: 3338–3344.
Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O'Donnell E et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res 2012; 18: 1611–1618.
Brune V, Tiacci E, Pfeil I, Doring C, Eckerle S, van Noesel CJM et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med 2008; 205: 2251–2268.
Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003; 198: 851–862.
Schwering I, Bräuninger A, Distler V, Jesdinsky J, Diehl V, Hansmann ML et al. Profiling of Hodgkin's lymphoma cell line L1236 and germinal center B cells: identification of Hodgkin's lymphoma-specific genes. Mol Med 2003; 9: 85–95.
Piva R, Agnelli L, Pellegrino E, Todoerti K, Grosso V, Tamagno I et al. Gene expression profiling uncovers molecular classifiers for the recognition of anaplastic large-cell lymphoma within peripheral T-cell neoplasms. J Clin Oncol 2010; 28: 1583–1590.
Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008; 322: 1097–1100.
Murphy TL, Tussiwand R, Murphy KM . Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol 2013; 13: 499–509.
Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 2012; 490: 502–507.
Logan MR, Jordan-Williams KL, Poston S, Liao J, Taparowsky EJ . Overexpression of Batf induces an apoptotic defect and an associated lymphoproliferative disorder in mice. Cell Death Dis 2012; 3: e310.
Weber K, Bartsch U, Stocking C, Fehse B . A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther 2008; 16: 698–706.
Tiacci E, Döring C, Brune V, van Noesel CJ, Klapper W, Mechtersheimer G et al. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood 2012; 120: 4609–4620.
Steidl C, Diepstra A, Lee T, Chan FC, Farinha P, Tan K et al. Gene expression profiling of microdissected Hodgkin Reed-Sternberg cells correlates with treatment outcome in classical Hodgkin lymphoma. Blood 2012; 120: 3530–3540.
Underhill GH, George D, Bremer EG, Kansas GS . Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood 2003; 101: 4013–4021.
Watanabe M, Ogawa Y, Ito K, Higashihara M, Kadin ME, Abraham LJ et al. AP-1 mediated relief of repressive activity of the CD30 promoter microsatellite in Hodgkin and Reed-Sternberg cells. Am J Pathol 2003; 163: 633–641.
Watanabe M, Sasaki M, Itoh K, Higashihara M, Umezawa K, Kadin ME et al. JunB induced by constitutive CD30-extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase signaling activates the CD30 promoter in anaplastic large cell lymphoma and reed-sternberg cells of Hodgkin lymphoma. Cancer Res 2005; 65: 7628–7634.
Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 2012; 490: 543–546.
Seitz V, Butzhammer P, Hirsch B, Hecht J, Gutgemann I, Ehlers A et al. Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS ONE 2011; 6: e26837.
Weilemann A, Grau M, Erdmann T, Merkel O, Sobhiafshar U, Anagnostopoulos I et al. Essential role of IRF4 and MYC signaling for survival of anaplastic large cell lymphoma. Blood 2015; 125: 124–132.
Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol 2012; 13: 1083–1091.
Iavarone C, Catania A, Marinissen MJ, Visconti R, Acunzo M, Tarantino C et al. The platelet-derived growth factor controls c-myc expression through a JNK- and AP-1-dependent signaling pathway. J Biol Chem 2003; 278: 50024–50030.
Chisholm KM, Bangs CD, Bacchi CE, Molina-Kirsch H, Cherry A, Natkunam Y . Expression profiles of MYC protein and MYC gene rearrangement in lymphomas. Am J Surg Pathol 2015; 39: 294–303.
Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell 2010; 18: 590–605.
Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015; 27: 516–532.
Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 2008; 111: 1515–1523.
Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008; 111: 3701–3713.
Skinnider BF, Elia AJ, Gascoyne RD, Patterson B, Trümper L, Kapp U et al. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2002; 99: 618–626.
Atsaves V, Lekakis L, Drakos E, Leventaki V, Ghaderi M, Baltatzis GE et al. The oncogenic JUNB/CD30 axis contributes to cell cycle deregulation in ALK+ anaplastic large cell lymphoma. Br J Haematol 2014; 167: 514–523.
Hartmann S, Martin-Subero JI, Gesk S, Husken J, Giefing M, Nagel I et al. Detection of genomic imbalances in microdissected Hodgkin and Reed-Sternberg cells of classical Hodgkin's lymphoma by array-based comparative genomic hybridization. Haematologica 2008; 93: 1318–1326.
Salipante SJ, Adey A, Thomas A, Lee C, Liu YJ, Kumar A et al. Recurrent somatic loss of TNFRSF14 in classical Hodgkin lymphoma. Genes Chromosomes Cancer 2016; 55: 278–287.
Feys T, Poppe B, De Preter K, Van Roy N, Verhasselt B, De Paepe P et al. A detailed inventory of DNA copy number alterations in four commonly used Hodgkin's lymphoma cell lines. Haematologica 2007; 92: 913–920.
Salaverria I, Bea S, Lopez-Guillermo A, Lespinet V, Pinyol M, Burkhardt B et al. Genomic profiling reveals different genetic aberrations in systemic ALK-positive and ALK-negative anaplastic large cell lymphomas. Br J Haematol 2008; 140: 516–526.
Liu C, Iqbal J, Teruya-Feldstein J, Shen Y, Dabrowska MJ, Dybkaer K et al. MicroRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood 2013; 122: 2083–2092.
Lamprecht B, Kreher S, Anagnostopoulos I, Johrens K, Monteleone G, Jundt F et al. Aberrant expression of the Th2 cytokine IL-21 in Hodgkin lymphoma cells regulates STAT3 signaling and attracts Treg cells via regulation of MIP-3alpha. Blood 2008; 112: 3339–3347.
Scheeren FA, Diehl SA, Smit LA, Beaumont T, Naspetti M, Bende RJ et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5; evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood 2008; 111: 4706–4715.
Gunawardana J, Chan FC, Telenius A, Woolcock B, Kridel R, Tan KL et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat Genet 2014; 46: 329–335.
Joos S, Küpper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res 2000; 60: 549–552.
Weniger MA, Melzner I, Menz CK, Wegener S, Bucur AJ, Dorsch K et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006; 25: 2679–2684.
Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med 2005; 11: 623–629.
Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene 2002; 21: 1038–1047.
Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011; 470: 115–119.
Rui L, Drennan AC, Ceribelli M, Zhu F, Wright GW, Huang DW et al. Epigenetic gene regulation by Janus kinase 1 in diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 2016; 113: E7260–E7267.
Kreher S, Bouhlel MA, Cauchy P, Lamprecht B, Li S, Grau M et al. Mapping of transcription factor motifs in active chromatin identifies IRF5 as key regulator in classical Hodgkin lymphoma. Proc Natl Acad Sci USA 2014; 111: E4513–E4522.
Carbone A, Gloghini A, Aldinucci D, Gattei V, Dalla-Favera R, Gaidano G . Expression pattern of MUM1/IRF4 in the spectrum of pathology of Hodgkin's disease. Br J Haematol 2002; 117: 366–372.
Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W et al. IRF4 addiction in multiple myeloma. Nature 2008; 454: 226–231.
Yang Y, Shaffer AL 3rd, Emre NC, Ceribelli M, Zhang M, Wright G et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 2012; 21: 723–737.
Shaffer AL, Staudt LM . The case of the missing c-Myc. Nat Immunol 2012; 13: 1029–1031.
Acknowledgements
We thank Kerstin Heise, Kristin Rosowski, Philip Abstoß and Klaus Lennartz for excellent technical assistance. Cells were sorted using a FACS ARIA III at the imaging facility Essen IMCES. This work was supported by grants from the Deutsche Forschungsgemeinschaft (KU1315/7-1, KU1315/10-1, GRK1431/2) and the BMBF through the International Cancer Genome Consortium for malignant lymphomas (01KU1002F). The MS experiments were supported by PURE (Protein Research Unit Ruhr within Europe, Ministry of Science, North Rhine-Westphalia, Germany).
Author contributions
AL, RK and MAW designed research; AL, SH, MS, TB, ALW and MAW performed research; TB, LK-H and BS contributed to analytic methods and tools; SH, JA and M-LH contributed to clinical samples; AL, SH, TB, BS, LK-H, M-LH, RK and MAW analyzed data; AL, RK and MAW wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Lollies, A., Hartmann, S., Schneider, M. et al. An oncogenic axis of STAT-mediated BATF3 upregulation causing MYC activity in classical Hodgkin lymphoma and anaplastic large cell lymphoma. Leukemia 32, 92–101 (2018). https://doi.org/10.1038/leu.2017.203
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.203
This article is cited by
-
siRNA-mediated downregulation of BATF3 diminished proliferation and induced apoptosis through downregulating c-Myc expression in chronic myelogenous leukemia cells
Molecular Biology Reports (2024)
-
Concise review: The heterogenous roles of BATF3 in cancer oncogenesis and dendritic cells and T cells differentiation and function considering the importance of BATF3-dependent dendritic cells
Immunogenetics (2024)
-
Challenges and new technologies in adoptive cell therapy
Journal of Hematology & Oncology (2023)
-
Transcriptional and epigenetic regulators of human CD8+ T cell function identified through orthogonal CRISPR screens
Nature Genetics (2023)
-
TET2 guards against unchecked BATF3-induced CAR T cell expansion
Nature (2023)