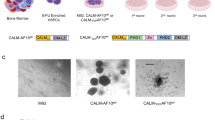
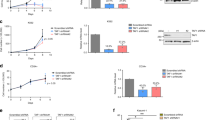
Abstract
The leukemogenic CALM-AF10 fusion protein is found in patients with immature acute myeloid and T-lymphoid malignancies. CALM-AF10 leukemias display abnormal H3K79 methylation and increased HOXA cluster gene transcription. Elevated expression of HOXA genes is critical for leukemia maintenance and progression; however, the precise mechanism by which CALM-AF10 alters HOXA gene expression is unclear. We previously determined that CALM contains a CRM1-dependent nuclear export signal (NES), which is both necessary and sufficient for CALM-AF10-mediated leukemogenesis. Here, we find that interaction of CALM-AF10 with the nuclear export receptor CRM1 is necessary for activating HOXA gene expression. We show that CRM1 localizes to HOXA loci where it recruits CALM-AF10, leading to transcriptional and epigenetic activation of HOXA genes. Genetic and pharmacological inhibition of the CALM–CRM1 interaction prevents CALM-AF10 enrichment at HOXA chromatin, resulting in immediate loss of transcription. These results provide a comprehensive mechanism by which the CALM-AF10 translocation activates the critical HOXA cluster genes. Furthermore, this report identifies a novel function of CRM1: the ability to bind chromatin and recruit the NES-containing CALM-AF10 transcription factor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Krumlauf R . Hox genes in vertebrate development. Cell 1994; 78: 191–201.
Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA 1994; 91: 12223–12227.
Pineault N, Helgason CD, Lawrence HJ, Humphries RK . Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol 2002; 30: 49–57.
Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia 1999; 13: 1993–1999.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286: 531–537.
Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol 1997; 17: 495–505.
Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G . Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J 1998; 17: 3714–3725.
Bohlander SK, Muschinsky V, Schrader K, Siebert R, Schlegelberger B, Harder L et al. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia 2000; 14: 93–99.
Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia 2005; 19: 1948–1957.
Caudell D, Zhang Z, Chung YJ, Aplan PD . Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res 2007; 67: 8022–8031.
Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y . Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol 2006; 8: 1017–1024.
Lin YH, Kakadia PM, Chen Y, Li YQ, Deshpande AJ, Buske C et al. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood 2009; 114: 651–658.
Conway AE, Scotland PB, Lavau CP, Wechsler DS . A CALM-derived nuclear export signal is essential for CALM-AF10-mediated leukemogenesis. Blood 2013; 121: 4758–4768.
Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167–178.
Chen L, Deshpande AJ, Banka D, Bernt KM, Dias S, Buske C et al. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia 2012; 27: 813–822.
Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011; 20: 53–65.
Fornerod M, Ohno M, Yoshida M, Mattaj IW . CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997; 90: 1051–1060.
Archangelo LF, Glasner J, Krause A, Bohlander SK . The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene 2006; 25: 4099–4109.
Morita S, Kojima T, Kitamura T . Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 2000; 7: 1063–1066.
Scotland PB, Heath JL, Conway AE, Porter NB, Armstrong MB, Walker JA et al. The PICALM protein plays a key role in iron homeostasis and cell proliferation. PLoS ONE 2012; 7: e44252.
Narita M, Shimizu K, Hayashi Y, Taki T, Taniwaki M, Hosoda F et al. Consistent detection of CALM-AF10 chimaeric transcripts in haematological malignancies with t(10;11)(p13;q14) and identification of novel transcripts. Br J Haematol 1999; 105: 928–937.
Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 2010; 17: 609–621.
Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 1999; 96: 9112–9117.
Sobell HM . Actinomycin and DNA transcription. Proc Natl Acad Sci USA 1985; 82: 5328–5331.
Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK . The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA 1996; 93: 4804–4809.
Yu W, Chory EJ, Wernimont AK, Tempel W, Scopton A, Federation A et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat Commun 2012; 3: 1288.
Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA . Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004; 117: 427–439.
Kalverda B, Pickersgill H, Shloma VV, Fornerod M . Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010; 140: 360–371.
Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW . Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010; 140: 372–383.
Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM . A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 1999; 18: 1660–1672.
McBride KM, McDonald C, Reich NC . Nuclear export signal located within the DNA-binding domain of the STAT1transcription factor. EMBO J 2000; 19: 6196–6206.
Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997; 390: 308–311.
Johnson C, Van Antwerp D, Hope TJ . An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IkappaBalpha. EMBO J 1999; 18: 6682–6693.
Baranek C, Sock E, Wegner M . The POU protein Oct-6 is a nucleocytoplasmic shuttling protein. Nucleic Acids Res 2005; 33: 6277–6286.
Rehberg S, Lischka P, Glaser G, Stamminger T, Wegner M, Rosorius O . Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation. Mol Cell Biol 2002; 22: 5826–5834.
Schmidt J, Braggio E, Kortuem KM, Egan JB, Zhu YX, Xin CS et al. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia 2013; 27: 2357–2365.
Takeda A, Sarma NJ, Abdul-Nabi AM, Yaseen NR . Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins. J Biol Chem 2010; 285: 16248–16257.
Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 2008; 111: 4668–4680.
Ghannam G, Takeda A, Camarata T, Moore MA, Viale A, Yaseen NR . The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J Biol Chem 2004; 279: 866–875.
Takeda A, Yaseen NR . Nucleoporins and nucleocytoplasmic transport in hematologic malignancies. Semin Cancer Biol 2014; 27: 3–10.
Newlands ES, Rustin GJ, Brampton MH . Phase I trial of elactocin. Br J Cancer 1996; 74: 648–649.
Etchin J, Sanda T, Mansour MR, Kentsis A, Montero J, Le BT et al. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol 2013; 161: 117–127.
Etchin J, Sun Q, Kentsis A, Farmer A, Zhang ZC, Sanda T et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia 2013; 27: 66–74.
Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 2012; 120: 1765–1773.
Acknowledgements
We appreciate the helpful critiques, comments and suggestions of Chi Dang and Rob Wechsler-Reya. We thank Jay Hess for providing the Hoxa9-LUC construct. This work was supported by NCI R01 CA 109281 (DSW), a V Foundation/Applebee’s Grant (DSW), a St Baldrick’s Research Grant (DSW) and a Hyundai Hope Grant (CPL).
Author Contributions
AEC and JMH designed and performed experiments, analyzed data and wrote the paper. DSW and CPL designed experiments and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Conway, A., Haldeman, J., Wechsler, D. et al. A critical role for CRM1 in regulating HOXA gene transcription in CALM-AF10 leukemias. Leukemia 29, 423–432 (2015). https://doi.org/10.1038/leu.2014.221
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.221
This article is cited by
-
MOZ/ENL complex is a recruiting factor of leukemic AF10 fusion proteins
Nature Communications (2023)
-
Fusion of the CRM1 nuclear export receptor to AF10 causes leukemia and transcriptional activation of HOXA genes
Leukemia (2021)
-
The role of the PZP domain of AF10 in acute leukemia driven by AF10 translocations
Nature Communications (2021)
-
Interaction with XPO1 is essential for SETBP1 to induce myeloid transformation
Leukemia (2019)
-
Polycomb repressive complex 2 facilitates the nuclear export of the influenza viral genome through the interaction with M1
Scientific Reports (2016)