Abstract
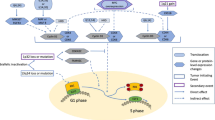
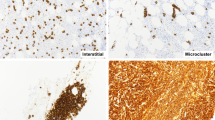
The molecular pathways implicated in multiple myeloma (MM) development are rather unknown. We studied epigenetic and DNA damage response (DDR) signals at selected model loci (N-ras, p53, d-globin) in bone marrow plasma cells and peripheral blood mononuclear cells (PBMCs) from patients with monoclonal gammopathy of undetermined significance (MGUS; n=20), smoldering/asymptomatic MM (SMM; n=29) and MM (n=18), as well as in healthy control-derived PBMCs (n=20). In both tissues analyzed, a progressive, significant increase in the looseness of local chromatin structure, gene expression levels and DNA repair efficiency from MGUS to SMM and finally to MM was observed (all P<0.002). Following ex vivo treatment with melphalan, a gradual suppression of the apoptotic pathway occurred in samples collected at different stages of myelomagenesis, with the severity and duration of the inhibition of RNA synthesis, p53 phosphorylation at serine15 and induction of apoptosis being higher in MGUS than SMM and lowest in MM patients (all P<0.0103). Interestingly, for all endpoints analyzed, a strong correlation between plasma cells and corresponding PBMCs was observed (all P<0.0003). We conclude that progressive changes in chromatin structure, transcriptional activity and DDR pathways during myelomagenesis occur in malignant plasma cells and that these changes are also reflected in PBMCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Anderson KC, Carrasco RD . Pathogenesis of myeloma. Annu Rev Pathol 2011; 6: 249–274.
Chng WJ, Glebov O, Bergsagel PL, Kuehl WM . Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol 2007; 20: 571–596.
Kyle RA, Rajkumar SV . Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma. Curr Hematol Malig Rep 2010; 5: 62–69.
Rajkumar SV, Gupta V, Fonseca R, Dispenzieri A, Gonsalves WI, Larson D et al. Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia 2013; 27: 1738–1744.
Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell 2006; 9: 313–325.
Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 2006; 108: 1733–1743.
Lopez-Corral L, Sarasquete ME, Bea S, Garcia-Sanz R, Mateos MV, Corchete LA et al. SNP-based mapping arrays reveal high genomic complexity in monoclonal gammopathies, from MGUS to myeloma status. Leukemia 2012; 26: 2521–2529.
Munshi NC, Avet-Loiseau H . Genomics in multiple myeloma. Clin Cancer Res 2011; 17: 1234–1242.
Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res 2004; 64: 1546–1558.
Lopez-Corral L, Mateos MV, Corchete LA, Sarasquete ME, de la Rubia J, de Arriba F et al. Genomic analysis of high-risk smoldering multiple myeloma. Haematologica 2012; 97: 1439–1443.
Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood 2002; 100: 1417–1424.
Lopez-Corral L, Gutierrez NC, Vidriales MB, Mateos MV, Rasillo A, Garcia-Sanz R et al. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res 2011; 17: 1692–1700.
Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH . DNA damage response and transcription. DNA Repair 2011; 10: 743–750.
Jasin M . Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 2002; 21: 8981–8993.
Clingen PH, Wu JY, Miller J, Mistry N, Chin F, Wynne P et al. Histone H2AX phosphorylation as a molecular pharmacological marker for DNA interstrand crosslink cancer chemotherapy. Biochem Pharmacol 2008; 76: 19–27.
Velangi MR, Matheson EC, Morgan GJ, Jackson GH, Taylor PR, Hall AG et al. DNA mismatch repair pathway defects in the pathogenesis and evolution of myeloma. Carcinogenesis 2004; 25: 1795–1803.
Sharma A, Heuck CJ, Fazzari MJ, Mehta J, Singhal S, Greally JM et al. DNA methylation alterations in multiple myeloma as a model for epigenetic changes in cancer. Wiley Interdisciplinary Reviews. Syst Biol Med 2010; 2: 654–669.
Walters DK, Wu X, Tschumper RC, Arendt BK, Huddleston PM, Henderson KJ et al. Evidence for ongoing DNA damage in multiple myeloma cells as revealed by constitutive phosphorylation of H2AX. Leukemia 2011; 25: 1344–1353.
Stefanou DT, Episkopou H, Kyrtopoulos SA, Bamias A, Gkotzamanidou M, Bamia C et al. Development and validation of a PCR-based assay for the selection of patients more likely to benefit from therapeutic treatment with alkylating drugs. Br J Clin Pharmacol 2012; 74: 842–853.
Souliotis VL, Dimopoulos MA, Sfikakis PP . Gene-specific formation and repair of DNA monoadducts and interstrand cross-links after therapeutic exposure to nitrogen mustards. Clin Cancer Res 2003; 9: 4465–4474.
Dimopoulos MA, Souliotis VL, Anagnostopoulos A, Papadimitriou C, Sfikakis PP . Extent of damage and repair in the p53 tumor-suppressor gene after treatment of myeloma patients with high-dose melphalan and autologous blood stem-cell transplantation is individualized and may predict clinical outcome. J Clin Oncol 2005; 23: 4381–4389.
Dimopoulos MA, Souliotis VL, Anagnostopoulos A, Bamia C, Pouli A, Baltadakis I et al. Melphalan-induced DNA damage in vitro as a predictor for clinical outcome in multiple myeloma. Haematologica 2007; 92: 1505–1512.
Salhia B, Baker A, Ahmann G, Auclair D, Fonseca R, Carpten J . DNA methylation analysis determines the high frequency of genic hypomethylation and low frequency of hypermethylation events in plasma cell tumors. Cancer Res 2010; 70: 6934–6944.
Chesi M, Bergsagel PL . Many multiple myelomas: making more of the molecular mayhem. Hematology (Am Soc Hematol Educ Program) 2011; 2011: 344–353.
Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 2011; 117: 211–220.
Morgan GJ, Kaiser MF . How to use new biology to guide therapy in multiple myeloma. Hematology 2012; 2012: 342–349.
Episkopou H, Kyrtopoulos SA, Sfikakis PP, Fousteri M, Dimopoulos MA, Mullenders LHF et al. Association between transcriptional activity, local chromatin structure and the efficiencies of both subpathways of nucleotide excision repair of melphalan adducts. Cancer Res 2009; 69: 4424–4433.
Souliotis VL, Dimopoulos MA, Episkopou HG, Kyrtopoulos SA, Sfikakis PP . Preferential in vivo DNA repair of melphalan-induced damage in human genes is greatly affected by the local chromatin structure. DNA Repair 2006; 5: 972–985.
Episkopou H, Kyrtopoulos SA, Sfikakis PP, Dimopoulos MA, Souliotis VL . The repair of melphalan-induced DNA adducts in the transcribed strand of active genes is subject to a strong polarity effect. Mutat Res 2011; 714: 78–87.
Matthews JN, Altman DG, Campbell MJ, Campbell MJ, Royston P . Analysis of serial measurements in medical research. BMJ 1990; 300: 230–235.
Ljungman M, Zhang F, Chen F, Rainbow AJ, McKay BC . Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene 1999; 18: 583–592.
Amano T, Nakamizo A, Mishra SK, Gumin J, Shinojima N, Sawaya R et al. Simultaneous phosphorylation of p53 at serine 15 and 20 induces apoptosis in human glioma cells by increasing expression of pro-apoptotic genes. J Neurooncol 2009; 92: 357–371.
Davies FE, Dring AM, Li C, Rawstron AC, Shammas MA, O'Connor SM et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood 2003; 102: 4504–4511.
Heuck CJ, Mehta J, Bhagat T, Gundabolu K, Yu Y, Khan S et al. Myeloma is characterized by stage-specific alterations in DNA methylation that occur early during myelomagenesis. J Immunol 2013; 190: 2966–2975.
Reinhardt HC, Schumacher B . The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet 2012; 28: 128–136.
Halazonetis TD, Gorgoulis VG, Bartek J . An oncogene-induced DNA damage model for cancer development. Science 2008; 319: 1352–1355.
Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005; 434: 907–913.
Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006; 444: 633–637.
Campisi J . Suppressing cancer: the importance of being senescent. Science 2005; 309: 886–887.
Rotunno M, Hu N, Su H, Wang C, Goldstein AM, Bergen AW et al. A gene expression signature from peripheral whole blood for stage I lung adenocarcinoma. Cancer Prev Res 2011; 4: 1599–1608.
Zander T, Hofmann A, Staratschek-Jox A, Classen S, Debey-Pascher S, Maisel D et al. Blood-based gene expression signatures in non-small cell lung cancer. Clin Cancer Res 2011; 17: 3360–3367.
Hagmar L, Brøgger A, Hansteen IL, Heim S, Högstedt B, Knudsen L et al. Cancer risk in humans predicted by increased levels of chromosomal damage. Cancer Res 1994; 54: 2919–2922.
Wu X, Amos CI, Zhu Y, Zhao H, Grossman BH, Shay JW et al. Telomere dysfunction: a potential cancer predisposition factor. J Nat Cancer Inst 2003; 95: 1211–1218.
Wu HC, John EM, Ferris JS, Keegan TH, Chung WK, Andrulis I et al. Global DNA methylation levels in girls with and without a family history of breast cancer. Epigenetics 2011; 6: 29–33.
Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology 2008; 134: 47–55.
Thiago LS, Perez-Andres M, Balanzategui A, Sarasquete ME, Paiva B, Jara-Acevedo M et al. Circulating clonotypic B-cells in multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica 2013; e-pub ahead of print 19 July doi:10.3324/haematol.2013.092817.
Bianchi G, Kyle RA, Larson DR, Witzig TE, Kumar S, Dispenzieri A et al. High levels of peripheral blood circulating plasma cells as a specific risk factor for progression of smoldering multiple myeloma. Leukemia 2013; 27: 680–685.
Matsui W, Borrello I, Mitsiades C . Autologous stem cell transplantation and multiple myeloma cancer stem cells. Biol Blood Marrow Transplant 2012; 18: S27–S32.
Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y et al. Characterization of clonogenic multiple myeloma cells. Blood 2004; 103: 2332–2336.
Acknowledgements
This work was partly supported by the ECNIS (Environmental Cancer, Nutrition and Individual Susceptibility) Network of Excellence of the European Union (contract no. 513943).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
MG performed the epigenetic and DNA damage response experiments, collected and analyzed the data, and wrote the manuscript; ET, PPS and SAK evaluated results and reviewed the manuscript; CB performed the statistical analysis of data and reviewed the manuscript; MAD was responsible for the selection and clinical evaluation of patients and reviewed the manuscript; and VLS designed and supervised the conducted research, performed the epigenetic and DNA damage response experiments, analyzed data, wrote the manuscript and approved the final version for submission.
Rights and permissions
About this article
Cite this article
Gkotzamanidou, M., Terpos, E., Bamia, C. et al. Progressive changes in chromatin structure and DNA damage response signals in bone marrow and peripheral blood during myelomagenesis. Leukemia 28, 1113–1121 (2014). https://doi.org/10.1038/leu.2013.284
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.284
Keywords
This article is cited by
-
miRNA-seq identification and clinical validation of CD138+ and circulating miR-25 in treatment response of multiple myeloma
Journal of Translational Medicine (2023)
-
Defective DNA repair and chromatin organization in patients with quiescent systemic lupus erythematosus
Arthritis Research & Therapy (2016)
-
Altered primary chromatin structures and their implications in cancer development
Cellular Oncology (2016)
-
Chromatin structure, transcriptional activity and DNA repair efficiency affect the outcome of chemotherapy in multiple myeloma
British Journal of Cancer (2014)