Abstract
Notch signaling is a key signaling pathway for cell proliferation and differentiation. Therefore, we formulated a working hypothesis that Notch signaling can be used to detect early osteoblastic differentiation of mesenchymal stromal cells. Changes in expression and distribution of Notch 1, 2, 3, and Delta1 in the cytoplasm and nuclei of rat liver-derived mesenchymal stromal cells differentiating into osteoblasts were investigated, together with the displacement of intracellular domains (ICDs) of the receptors. In addition, an oligonucleotide microarray was used to determine the expression of genes known to be linked to selected signaling pathways. Statistically significant changes in the number of cells expressing Notch1, Notch2, and Delta1, but not Notch3, and their activated forms were detected within 24 h of culture under osteogenic conditions. Although the number of cells expressing Notch3 remained unchanged, the number of cells with the activated receptor was significantly elevated. The number of cells positive for Notch3 was higher than that for the other Notch receptors even after 48 h of differentiation; however, a smaller fraction of cells contained activated Notch3. Culture mineralization was detected on day 4 of differentiation, and all analyzed receptors were present in the cells at that time, but only Delta1 was activated in twice as many cells than that before differentiation. Thus, the three analyzed receptors and ligand can serve as markers of very early stages of osteogenesis in stromal cells. These early changes in activation of the Notch signaling pathway were correlated with the transcription of several genes linked to osteogenesis, such as Bmps, Mmps, and Egfr, and with the regulation of cell cycle and apoptosis.
Similar content being viewed by others
Main
The field of regenerative medicine is witnessing the use of pluripotent stem cells as potential candidates for engineering various tissues or their fragments, including bone, for surgical treatment.1, 2 Stem cells have numerous advantages, but they are most useful for organ and tissue regeneration due to their high potential for proliferation and differentiation.3, 4 However, there are also some risks related to these two features, such as the potential of neoplastic transformation following transplantation.3, 5 Therefore, different options for the use of stem cells in regeneration have been explored, including the use of defined progenitors of specific cell lineages.6, 7 However, orchestrating the genetic programs in stem cells for their differentiation to specific cell types requires perfect understanding of mechanisms and signal transduction pathways governing these processes.
For the treatment of bone injury or bone disorders and diseases, two major cell types are required. These include osteoblasts that build the new bones at the site of injury and osteoclasts that participate in bone turnover at the orthopedic interfaces, for example, implant–bone interfaces.8, 9 Here we have investigated the cellular changes occurring in adult mesenchymal stromal cells during the early stages of differentiation to osteoblasts.
The cells investigated here were mesenchymal stromal cells from livers of adult rats that we had previously characterized and described.10 These cells have the capabilities of transformation into non-hepatic cell lineages.10 They also differentially express Notch1, 2, and 3 receptors and Dll1 depending on their age and physiological stage.10 However, the exact signaling mechanisms involved in the differentiation of liver-derived mesenchymal stromal cells into osteoblasts need to be investigated further.
A possible regulatory pathway for differentiation during different stages of fetal development and in the postnatal life of organisms involves Notch receptors.11, 12 The Notch signal transduction leads to differentiation of progenitor cells into different cell lineages.13 This signaling pathway is also involved in the regulation of cell proliferation and response to hypoxia, as well as in the induction of apoptosis and formation of stem cell niche.14, 15 In mammals, including humans, four genes encoding Notch1, 2, 3, and 4 receptors have been identified.14 In addition, five genes encoding Notch receptor ligands have been reported, namely, Jagged1, 2, and 3 and Delta-like1 and 2 (Dll1 and Dll2).14, 15, 16 Among Notch-related pathways, the most extensively investigated is the one involving Notch1 and Dll1. Although the number of reports on the role of other members of the Notch family has recently increased, the relationship between these receptors and ligands and the mechanisms of regulation of the Notch signaling pathways are still poorly understood. Intracellular regulation of Notch signaling occurs near the cell membrane and then in the nucleus. This multilevel regulation directly links the regulatory errors in Notch signaling to several human disorders, such as developmental disorders, cardiovascular disorders and cancer.17, 18, 19 The constitutively active Notch signaling pathway usually acts in a pro-oncogenic manner with the exception of a case reported by Koch and Radtke,20 where it was shown to suppress tumorigenesis.
The Notch signaling pathway acts over short distances. It involves interactions between neighboring cells, leading to changes in the expression of Notch-related genes. An extracellular Notch/ligand interaction first triggers proteolytic cleavage of the Notch receptor and subsequent release of its extracellular domain. Further signaling is facilitated by the cleavage of intracellular polypeptide chain, called Notch IC domain (ICD). Following the second cleavage, Notch ICD is translocated to the nucleus.21 In the nucleus, the domain interacts with DNA-binding transcription factors, such as CBF-1, Su(H), and Lag-1 (CSL) that are the nuclear effectors of the Notch signaling pathway.22 Thus, the cellular location of Notch ICD can be monitored as an indicator of pathway activation. In the nucleus, the Notch ICD, together with the transcription co-activator Mastermind, forms a complex with the CSL, which subsequently activates the transcription of Notch signaling-related genes.23
Numerous reports from studies involving the loss- and gain-of-function for Notch signaling pathway partners indicated that during early stages of osteogenesis in vivo, Notch maintains mesenchymal stromal cells in an undifferentiated stage and decreases osteoblast differentiation.12 Therefore, here, we have investigated the expression of four Notch receptor-encoding genes and two of their ligands, Dll1 and Jagged2, in liver-derived mesenchymal stromal cells subjected to differentiation into osteoblasts in vitro. In addition, we aimed to determine the Notch receptor that is activated at very early stages of stem cell differentiation into osteoblasts by monitoring Notch1, Notch2, Notch3, and Dll1 at both mRNA and protein levels. Subsequently, the intracellular translocation of Notch1, 2, and 3 ICDs was monitored.
Materials and methods
Isolation of Mesenchymal Stromal cells from Rat Livers
Approximately 2 g of liver tissues from healthy, 4-month-old, Sprague-Dawley rats was excised and extensively washed in PBS. Prior to organ collection, the animals were treated according to the procedure accepted by the Local Ethical Commission for Research Animal Use, No. 40/2007. Subsequently, each piece of liver tissue was separately minced and incubated in alpha-MEM supplemented with collagenase (2 mg/ml) at 37 °C for 60 min. Subsequently, the collagenase-treated samples were trypsinized (10 mg/ml) for 45 min. After the digestion was complete, each sample was suspended in 10 ml alpha-MEM, and the cells were recovered by centrifugation at 500 g for 10 min. This step was repeated twice. The cell pellet was suspended in 10 ml of alpha-MEM and centrifuged again to remove blood.
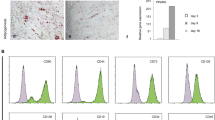
The cells were cultured until they reached ~80% confluence and then detached with the use of trypsin/EDTA (Sigma). The cells were recovered by centrifugation at 500 g for 10 min and characterized by fluorescence-assisted flow cytometry to detect cell surface antigens such as CD34, CD45, CXCR4, and lineage αβ T-cell receptor (Lin). Fluorescently labeled antibodies against CD34 (Santa Cruz Biotechnology, USA), CD45 (BioLegend), CXCR4 (BD Biosciences Pharmingen), and Lin (FITC anti-rat αβ T-cell receptor (R73), BD Biosciences Pharmingen) were used. The cells of interest were characterized by the presence of markers CXCR4 and CD34 and the lack of CD45 and Lin (Figure 1a). These cells were considered liver-derived mesenchymal stromal cells (MSCs).
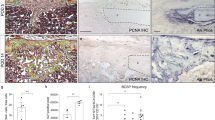
Characterization of liver-derived mesenchymal stromal cells by phenotype and expression and localization of Notch receptors and ligands in undifferentiated MSCs and during their differentiation to osteoblasts. (a) Detection of cell surface markers (CD34 and CXCR4) specific for pluripotent rodent MSCs in undifferentiated liver-derived MSCs by flow cytometry. (b) Detection of mRNAs encoding the four Notch receptors and their three ligands in the undifferentiated liver-derived MSCs. Lanes: M—DNA ladder; Notch 1, 2, 3, 4—products reflecting Notch1, 2, 3, and 4, respectively, Ligand, D1, J1, J2—products reflecting DLL1, Jagged1 and 2, respectively. The numbers under the bands indicate the expected length of the PCR product. The question mark denotes an unspecific product obtained for Jagged2. (c) Immunodetection and location of receptors: Notch1, 2, and 3 and ligand Dll1 in liver-derived MSCs differentiating to osteoblasts. The lowest panels show representative microphotographs of specimens with cells cultured under osteoblastic differentiation conditions stained with Alizarin Red S to determine the content of calcium phosphate (Ca2+). N.A.—indicates that Alizarin Red S staining was not performed for this time point. Arrows—cells with the detected antigen in the nucleus or in both the nucleus and cytoplasm. Arrowheads—cells with the detected antigen exclusively in the cytoplasm. Scale bars indicate 100 μm. MSCs, mesenchymal stromal cells.
Culture of Liver-Derived MSCs
Cells were suspended in 10 ml of MesenCult specialty medium recommended for the culture of rodent mesenchymal stromal cells (StemCell Technologies, Canada) and maintained for expansion in a Petri dishes in a humidified atmosphere with 95% air and 5% CO2 at 37 °C. At ~80% confluence, the cells were dissociated from the culture surface with a trypsin/EDTA mixture and split for further expansion in larger culture vessels that were double in size. Cells for immunostaining assays were plated in chamber glass slides (Lab-Tek Chamber Slide, NUNC, USA) at 2 × 104/1.8 cm2 and cultured in MesenCult medium for 24 h. Subsequently, the medium was replaced with the osteoblast differentiation medium or control culture medium, and the cells were cultured for 0, 1, 2, 3, 4, 6, 8, and 10 days. The media were changed every other day.
Differentiation of Liver-Derived MSCs
MesenCult medium was replaced with specialty differentiation media for osteoblasts (StemCell Technologies). The cultures were maintained for the indicated times in the differentiating medium. For differentiation of liver-derived MSCs to osteoblast, β-glycerophosphate (5 mM) with ascorbic acid was added from the beginning of differentiation and, the medium was replaced every other day.
Evaluation of Expression of Notch Receptors and Dll1 by PCR in Control and Differentiating Liver-Derived MSCs
Total RNA was extracted from cells using RNeasy Mini kit (Qiagen, Germany). The concentrations of mRNA were determined spectrophotometrically, and ~200 ng of total mRNA was used for reverse transcription. Single-stranded cDNA was obtained using Enhanced AMV Reverse Transcriptase (Sigma) and primed with Anchored Oligo (dT)23. Subsequently, the RT-PCR product was subjected to PCR using Taq polymerase using specific primers listed in Table 1. The following conditions were applied for PCR. PCR was carried out using 5 μl of the RT products and Accu Taq polymerase (Sigma) in a final volume of 50 μl. The samples were denatured at 95 °C for 30 s, annealed at 56 or 70 °C for 30 s and extended at 72 °C for 1 min. The cycle of incubations was repeated 40 times. The PCR products were identified after electrophoresis in 1.2% agarose gels, and the obtained products were verified by DNA sequencing using ABI Prism 310 DNA Analyser (Applied Biosystems).
Immunodetection of Notch Receptors and Ligand in Liver-Derived MSCs Following Differentiation to Osteoblasts
After incubation in osteogenic medium, cells were fixed for 20 min with 4% paraformaldehyde and subjected to detection with appropriate antibodies. The cells were stained with antibodies against ICDs for Notch1, 2, and 3 and for Dll1 (at dilution 1:50). Since the antibodies detected the ICDs, it was possible to discriminate the staining in the cytoplasm due to newly synthetized or inactive form of Notch from ICDs that had translocated in the nuclei upon activation following the second cleavage of Notch receptors. The binding of each ICD with its specific antibody was visualized by the detection of the corresponding secondary antibody. The secondary antibodies against Notch1 ICD and Dll1, which were conjugated with horseradish peroxidase, were visualized from the brown color of reaction product using DAB. The secondary antibodies against ICD Notch2 and 3 ICDs, which were conjugated with alkaline phosphatase (AP), were detected from the blue color of the reaction product using BCIP/NBT.
To determine the ratio of positive cells to negative cells for specific Notch ICDs or for Dll1, the mean values were calculated based on the cell number by microscopic observation. Cells were counted in triplicate, with a total of 1000 cells per count. The cells with ICDs in the cytoplasm and nuclei were considered to harbor activated Notch pathways. The cells positive for Dll1 were considered to be potentially ready to interact with the cells positive for Notch receptors.
Detection of Osteogenic Markers in Differentiating Liver-Derived MSCs
Cells that had differentiated to osteoblasts were identified by the detection of hydroxyapatites in culture by staining with Alizarin Red S (AR-S). The amount of deposited calcium phosphate was also assayed by a method described previously.10 Briefly, at particular time points during osteoblastic differentiation, the medium was removed, and the cells were washed with PBS followed by fixation with ice-cold 70% ethanol for at least 1 h. Following fixation, ethanol was removed, and the fixed cells were rinsed with deionized H2O and stained with 40 mM AR-S (pH 4.2) for 10 min at room temperature. The cells were then rinsed five times with H2O, followed by washing for 15 min with PBS to reduce unspecific AR-S binding. The samples were photographed and immediately assayed. After the AR-S dye bound to minerals, it was solubilized in 10% cetylpyridinium chloride (CPC) in 10 mM sodium phosphate, pH 7.0, for 15 min at 25 °C. The recovered AR-S was diluted 10 times in 10% CPC solution, and the concentration of AR-S was determined by measuring the absorbance of the sample at 562 nm and calculating the value using the formula obtained from the AR-S standard curve.
Determination of Cell Numbers
The number of cells in culture was determined using the metabolism of alamarBlue, which is a dye containing resazurin as the active component.24, 25 The alamarBlue assay is based on oxidation-reduction processes occurring in cells. The alamarBlue indicator is taken up by the cells and reduced at a rate dependent on the growth and metabolic activity of the cells. The reduced form of the dye is red and fluorescent, which was measured using a Fluoroscan plate reader. Fluorescence was monitored at 560 nm excitation wavelength and 590 nm emission wavelength. In this study, the alamarBlue reagent was added to the culture medium at a final concentration 10% four hours prior to the assay. The number of cells was calculated according to linear standard curves prepared separately for rat liver-derived mesenchymal stromal cells and for control osteoblasts (hFBO1.19 ATCC CRL-11372) at densities ranging from 4000 to 80 000 cells per well of 24-well plates. Fluorescence was measured using a Fluoroscan plate reader Victor X5 (Perkin Elmer), and based on the change in fluorescence reading with cell density, a linear function was calculated for both types of cells.
Gene Expression Profiling
A membrane-type oligonucleotide microarray (Rat Signalling Pathway Detection PCR Array; SABiosciences, Qiagen, USA/Germany) comprising sequences representing 113 receptor genes, 7 housekeeping genes, 2 detection controls, and 6 blank and artificial sequences was used for the analysis of gene expression profiles. Total mRNA was isolated from undifferentiated liver-derived MSCs cultured for 1, 2, or 8 days in proliferating medium and from stromal cells subjected to differentiation for 1, 2, or 8 days. To purify the mRNA, an ArrayGrade Total RNA Isolation Kit supplied by SABiosciences was used. The protocol for oligo microarray processing was supplied by the manufacturer, and the procedure recommended by the kit supplier was strictly followed. The genes were grouped into 20 groups of known signal transduction pathways (Supplementary Table S1). Some genes in the array are common for more than one group; therefore, the number of genes on the array and their total number in functional groups differed.
Statistical analysis
The results of quantitative assays are reported as the means±standard deviations (s.d.). Statistical significance of the differences for each Notch receptor (Notch1, 2, and 3) and Dll1 ligand for the same time point and same receptor and ligand during the differentiation process was calculated using parametric one-way ANOVA for normal distribution, followed by the Tukey–Kramer test. In case of non-parametric distribution, Kruskal–Wallis range test was applied, followed by the Steel–Dwass test. Prior to the above analyses, normal distribution was verified using D’Agostino–Pearson test.26 The homogeneity of variance was examined by Bartlett’s test. The accepted level of statistical significance was P<0.05. Statistical analyses were performed using Merlin Statistics Software27 and KyPlot, and the details are included in Supplementary Materials.
Results
Cultured Liver-Derived MSCs Were Not Homogenous
The cells obtained from rat livers under conditions optimized for culturing mesenchymal stromal cells revealed the presence of subpopulations characterized by different combinations of cell surface markers known as stem cell markers (Figure 1a). Analysis of the results from fluorescence-assisted flow cytometry assay revealed the presence of ~21% of cells positive for the markers CXCR4 and CD34, which are markers specific for undifferentiated and uncommitted stem cells (shaded boxes in Table 2). Cells positive for only one of these two markers alone or in combination with different markers accounted for ~42.3% of the parental pool. Cells positive for the hematopoietic markers CD45 and Lin accounted for 21.2%, whereas 6.6% of the parental pool was composed of cells negative for these two markers. The cells with only one of the two stem cell markers, ie, CD34 or CXCR4, detected in combination with one of the two hematopoietic markers represented ~21% each of the total cells (Table 2). Therefore, we concluded that the tested cells were a mixture of stromal cells containing progenitors of various cell lineages. As the cells were attached to the culture surface under conditions preferential for mesenchymal cells, we did not sort them further into homogeneous populations. Therefore, we accepted that all the cells were not osteoblast precursors and thus may not differentiate to osteoblasts under conditions used in this study.
Genes Encoding Notch Receptors and Their Ligands are Differentially Expressed in Liver-Derived MSCs
Qualitative analysis of the PCR products with specific primers for Notch1, 2, 3, and 4 as well as for their ligands, Dll1, and Jagged1 and 2, revealed their differential expression patterns in the liver-derived MSCs (lanes marked: Notch, 1, 2, 3, and 4 in Figure 1b). Particularly, Notch4 was undetectable in these cells. In addition, the expression of Dll1 but not that of Jagged1 (and only an unspecific product for Jagged2 marked with ‘?’ in Figure 1b) could be detected in the liver-derived MSCs (lanes marked: Ligand, D1, J1, and J2 in Figure 1b).
Liver-Derived MSCs Expressed Three Notch Receptors and the Ligand Dll1
Detection with specific antibodies against ICDs of Notch1, 2, and 3 receptors and Dll1 ligand in liver-derived MSCs revealed the presence of receptors and ligand both prior to and following culture in the osteoblast differentiating medium regardless of the incubation time (Figure 1c). Notch4 and Jagged1 and 2 were undetectable at both mRNA and protein levels. The detected ICDs differed in terms of their intracellular location and their corresponding type of Notch ligands. However, not all the cells revealed the presence of these antigens. Some cells did not react at all with particular antibodies, whereas some cells revealed the presence of a particular antigen exclusively in the nucleus or cytoplasm (indicated by arrows or arrowheads, respectively, in Figure 1c). Finally, in some cells, the detected antigens were present both in the nucleus and cytoplasm (Figure 1c). However, we cannot exclude that the possibility that cells not expressing a particular Notch receptor or ligand expressed a different Notch receptor, as multiple immunostainings were not performed due to the lack of differently tagged primary antibodies when this study was performed, as well as the lack of suitable secondary antibodies conjugated with distinct tags or enzymes.
Quantitative Changes in Cell Numbers with Active and Inactive Notch Pathways
The number of cells expressing Notch1, 2, and 3 receptors and Dll1 ligand at different time point during differentiation is presented in Supplementary Materials (Supplementary Tables S2–S4). Analyses of the number of cells with the presence of Notch1, 2, and 3 receptors or Dll1 ligand revealed the number of cells with Notch1 receptor had doubled by the first day of differentiation (P<0.05) (Supplementary Table S2). In addition, the number of cells expressing Notch2 receptor and Dll1 ligand increased by two-fold (P<0.001) and three-fold (P<0.001), respectively (Supplementary Table S2). The number of cells containing these antigens in the nuclei (filled boxes in Figure 2a) increased 4, 10, and 11 times (P<0.001) (Supplementary Table S3), indicating that there was an activation of the Notch signal transduction. Although, the number of cells expressing Notch3 receptor was almost unchanged on day 1 of differentiation (Supplementary Table S2), the number of cells containing Notch3 ICD in the nuclei (filled boxes in Figure 2a) was elevated by 11-fold (P<0.001) (Supplementary Table S3).
Fractions of cells with activated Notch signaling pathway during early differentiation of liver-derived MSCs to osteoblasts and detection of minerals in cultures. (a) Quantitation of cells expressing Notch1, 2, and 3 (N1, N2, and N3) and Dll1 (D) presented as ratio of cells that express the detected antigen to cells that do not (Supplementary Tables S2–S4). Distribution of Notch intracellular domains and Dll1 between the cytoplasm (open boxes) and nuclei (filled boxes) presented as percentage in the fraction of cells expressing the detected antigen. (b) Expression patterns of Notch1, 2, and 3 and Dll1 following agarose gel electrophoresis of PCR products with the use of primers listed in Table 1. The curve in the graph represents changes in mineral content (right axis) in the cultures during osteoblastic differentiation. Symbols: M—DNA ladder, Notch: 1—Notch1, 2—Notch2, 3—Notch3, D—Dll1 ligand. The indicated standard deviation on the histograms represent the ratios of the number of cells that are positively stained for the receptors and ligand to the number of cells that are negatively stained. MSCs, mesenchymal stromal cells.
On day 2 of differentiation, the number of cells expressing Notch1, 2, and 3 receptors and Dll1 ligand returned to the level of the control, but the number of cells expressing Notch3, remained unchanged (Supplementary Table S2). In all cases, the number of cells with ICDs of the factors in the nuclei returned to control values (Figure 2a,Supplementary Table S3). On days 3 and 4 of differentiation, the receptors could only be detected in a relatively small fraction of cells. However, on day 4 of differentiation, the number of cells expressing the ligand was again comparable to the number on day 1 of differentiation. In addition, on day 4, the first signs of mineralization could be detected (lowest panel in Figure 1c,Supplementary Table S2 and curve in Figure 2a). On day 6 of differentiation, the number of cells with Notch1 receptors, both cytoplasmic and nuclear, returned to the values observed on day 1. However, the number of cells with the other analyzed receptors and ligand were lower than that in the control cells (Notch2—P<0.001, Notch3—P<0.05, Dll1—P<0.001, and in the nuclei—P<0.01) (Figure 2a, Supplementary Tables S2 and S3). The number of cells with ICDs of Dll1 in the cytoplasm was unchanged in comparison to that of control cells only on day 4. On day 8, when the production of minerals has begun to peak (Figure 2a, Supplementary Table S2), the number of cells with all analyzed receptors and the ligand was elevated once again. However, the increase was statistically significant only for Notch2, Notch3, and Dll1 (P<0.001 for all) (Supplementary Table S4).
The results obtained from the immunodetection of Notch receptors and ligand Dll1 were correlated with the pattern of expression obtained using agarose gel electrophoresis of the PCR products (Figure 2b). Although, PCR is not quantitative, the weak signal for Notch3 in the total pool of cells following culture in MesenCult and under osteoblastic conditions could have resulted from the lower efficiency of PCR.
Liver-Derived MSCs Varied in Gene Expression Patterns During Early Stages of Their Differentiation to Osteoblasts
Statistically significant differences in gene expression levels were detected for 12 genes. On day 1 of cell culture in the osteogenic medium, seven genes displayed differential expression at statistically significant levels. However, none of these genes could be regarded as specific markers of osteogenesis (Figure 3, Supplementary Table S1). The seven genes that were differentially expressed in the stromal cells on day 1 still showed this trend on day 2, but the difference was not statistically significant except for Pten (Phosphatase and tensin homolog). Expression of Pten was significantly higher in cells cultured under osteogenic conditions. Pten is postulated to have a putative role in the regulation of phosphoinositide 3-kinase- and Akt-mediated inhibition of apoptosis. After 8 days of culture in the osteogenic medium, 10 genes were expressed at elevated levels and the differences were statistically significant. Five of these genes showed statistically significant differences in expression by day 1. However, four of these genes did not differ in their levels of expression. The gene Bmp2 was the only one whose expression was not detected after 1 and 2 days of differentiation, but its expression was significantly elevated in osteoblasts on day 8 (Figure 3). In addition, Bmp4, whose encoded product is known to interact with Bmp2 to form a heterodimer, displayed a trend toward elevated expression on day 8 of culture in the osteogenic medium (Figure 3).
Correlation analysis of expression patterns of genes related to signal transduction pathways using oligonucleotides microarrays on cells cultured in proliferating or differentiating culture medium with increasing duration of culture. Gene names and their reference numbers are provided in Supplementary Table S1. The lines on the graphs mark the range of differences in expression that is not statistically significant. Genes located outside the marked area differed in expression, and the difference was statistically significant.
Discussion
To regulate the process of bone formation by newly developed strategies, it is necessary to understand the mechanisms involved in the process of osteogenesis and bone repair. Therefore, here, we have investigated the Notch signaling pathway, which is the key regulator of cell fate decisions in prenatal skeletal development and is active during adult tissue renewal. Previous studies by others have revealed that attenuated Notch signaling enhances osteoclastogenesis and bone resorption in vitro and in vivo by a combination of molecular mechanisms.12 Deletion of NOTCH1, 2, and 3 in bone marrow macrophages, which are osteoclast precursors, directly promotes their commitment to the osteoclast lineage mediated by macrophage colony-stimulating factor and RANKL, leading to enhanced differentiation in response to low doses of either cytokine.12 Presentation of the NOTCH ligand, JAGGED1, abrogates the capacity of wild-type bone marrow macrophages to differentiate into osteoclasts. Altogether, these data demonstrate that NOTCH suppresses osteoclastogenesis via ligand-mediated receptor activation. Although NOTCH1 and NOTCH3 collaborate in regulating osteoclast formation, NOTCH1 is the dominant paralog. In addition, NOTCH1 deficiency indirectly promotes osteoclastogenesis by enhancing the ability of osteoblast lineage cells to stimulate osteoclastogenesis by decreasing the osteoprotegerin (Opg)/RANKL expression ratio. Thus, NOTCH1 acts as an inhibitor of bone resorption, exerting its effect both directly on osteoclast precursors and indirectly via osteoblast lineage cells.
In this study, we showed that mesenchymal stromal cells isolated from livers of young rats are a mixed population of adherent cells positive for markers that are specific for nonhematopoietic stem cells. In addition, the cells lack markers that are specific for non-adherent hematopoietic precursors. Previously, we reported that the population was also positive for Thy1 and CD34. CD34 is considered by some investigators to be a specific marker for hematopoietic precursors. On the other hand, a large number of reports indicates that CD34 is a marker that is present on both hematopoietic and non-hematopoietic precursors isolated from various tissues1, 28, 29 including livers in rodents.30 Thus, in this study, we further characterized a set of markers specific for mesenchymal stromal cells, indicating that there is a population of multipotent cells giving rise to progenitors of different cell types, including osteoblasts. Analysis of the flow cytometry results indicated that the parental pool of liver-derived mesenchymal stromal cells was a mixture of cells at different stages of partial commitment. The cells regarded as pluripotent stem cells expressing a combination of surface antigens CD45−/Lin−/CD34+/CXCR4+ was a minor cell fraction in the parental pool studied here. The presence of a population of cells positive for CD45 indicated that some cells had already acquired a hematopoietic fate and differentiated into hematopoietic progenitors. Thus, the cells were no longer multipotent stem cells. This mixture of pre-committed cells, prior to culture under osteogenic conditions, could explain the observation that not all cells expressed the same phenotype represented by the expression of Notch receptors and the Dll1 ligand. Another possible explanation of this phenomenon could be that the culture was not synchronized in cell cycle, metabolic activity, or cell-type commitment.
The results from the detection of gene expression of Notch receptors Dll1 and Jagged2 revealed that in the liver-derived MSCs, only the expression of Notch1, 2, and 3 and Dll1 was detected. Under the conditions examined in this study, the expression of Notch4 and Jagged1 and 2 was not detected. Therefore, only these four players of the Notch signaling pathway were further investigated over the course of osteogenic differentiation. Although the Notch3 receptor was present in more than 50% of the cells prior to the culture under osteogenic conditions, it was localized mostly in the cytoplasm; therefore, we speculate that Notch3 was in an inactive state. On day 1 of culture in the osteogenic medium, the presence of the Notch1 receptor, Dll1 ligand, and Notch2 receptor could be detected in 25%, 30% and ~ 50% of the cells, respectively. The number of cells with Notch3 receptor was lower than that prior to differentiation, but the other two Notch receptors and Dll1 were detected in the nuclei of more than 70% and more than 80% of cells, respectively. These results indicate that the activation of the three analyzed receptors is important for the initiation of osteogenic differentiation of liver-derived MSCs. The Notch3 receptor likely has a role over a longer period of time for long osteogenic differentiation, since the number of cells expressing this receptor was elevated on day 2 of differentiation.
The analysis of gene expression profiles during osteoblastic differentiation revealed a group of genes that were upregulated over the time of differentiation. Elevated expression of Pten during early osteogenesis could be linked to Notch signaling. Pten plays central role as a tumor suppressor and is best characterized for its role as a lipid phosphatase that acts to oppose the activation of the PI3K/Akt pathway.31, 32 It is known that Pten is regulated by the Notch1 signaling pathway.33, 34 It has also been shown that Pten is regulated transcriptionally by the Notch1 signaling pathway through the transcription factor Cbf1.35 The concentration of Pten is also increased by Bmp2.36 Bmp2 treatment in MCF-7 cells has been shown to cause a decrease in cellular proliferation, resulting in elevated levels of Pten due to its decreased association with proteins in the ubiquitin degradation pathway. Activation of Bmp type-1 receptor has been recently demonstrated to stimulate targeted degradation of Smad1.37 Pten expression is regulated at the transcriptional level by a set of transcriptional factors, including Tp53, and at the posttranscriptional level by protein localization, modification, and degradation.38 Although both Pten and Tp53 genes are frequently mutated in a variety of human cancers, their mutations are usually mutually exclusive.39 Recent evidence suggests that Pten expression is activated by Tp5340 and that they can physically interact.41 It was also shown that Pten upregulates its own expression by stabilizing Tp53 through a phosphatase independent mechanism.42 In our study, expression of both Tp53 and Pten was significantly elevated on day 8 of osteoblastic differentiation of liver-derived mesenchymal stromal cells.
The other genes that were upregulated on day 8 of osteogenesis such as Hspcal3 (Heat shock protein 90 kDa alpha (cytosolic), class A member 2—Hsp90AA2) and Hspb1 (Heat shock 27 kDa protein 1) are known as chaperones for various proteins. Cebpb is CCAAT/enhancer binding protein beta (C/Ebp), which is a bZip transcription factor that binds as a homodimer to certain DNA regulatory regions or as a dimer with other regulatory proteins. This protein can bind the promoter and upstream element and stimulate the expression of collagen type I gene, which is a major protein in the bone matrix.
Rbp2 is an abundant protein present in the small intestinal epithelium. It is thought to participate in the uptake and/or intracellular metabolism of vitamin A, which is a fat-soluble vitamin necessary for growth, reproduction and differentiation. Bax, which was found to be statistically significantly upregulated in our study, is linked to proapoptotic regulation via the mitochondrial pathway. In our study, the expression of Fos was also statistically significantly up-regulated on day 8. This gene encodes a leucine zipper protein, which can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1 to regulate cell proliferation, differentiation, and transformation. The last gene that was detected to be upregulated by the array during osteogenic differentiation of liver-derived MSCs was epidermal growth factor receptor (Egfr). The protein product of this gene is a transmembrane glycoprotein receptor, which upon binding of epidermal growth factor, dimerizes and leads to the induction of cell proliferation.
In conclusion, we have determined that for the osteogenic differentiation of liver-derived mesenchymal stromal cells, a coordinated induction of the expression of Notch1, 2, and 3 and Dll1 genes is critical. The mechanism for the role of Notch1 during early stages of osteogenic differentiation might involve known regulators of the cell cycle and apoptosis, namely, Pten/Tp53/Rbp2 and Bax, respectively, and factors involved in proliferation and differentiation, namely, Egfr and BMP2, respectively. Finally, chaperones such as Hspb1 and Hspcal3 were also expressed at higher levels on day 8 of osteoblastic differentiation. The expression of Notch1 is upregulated by Tp53, but signaling through the Notch1 receptor downregulates the expression of Tp53.43, 44 Thus, Tp53 could be a direct link between Notch1 signaling and cell cycle regulation in slowing down the proliferation of differentiating cells.
In summary, we have identified differential expression and activation of the three Notch receptors and their ligand Dll1 during early osteogenic differentiation accompanied by significant differences in gene expression profiles during the early stages of stem cell differentiation toward osteoblasts in comparison with undifferentiated stem cells.
References
Li SC, Acevedo J, Wang L et al, Stem cell engineering for treatment of heart diseases: potentials and challenges. Cell Biol Int 2009; 33: 255–267.
Ko EK, Jeong SI, Rim NG et al, In vitro osteogenic differentiation of human mesenchymal stem cells and in vivo bone formation in composite nanofiber meshes. Tissue Eng Part A 2008; 14: 2105–2119.
Dumas A, Moreau MF, Ghérardi RK et al, Bone grafts cultured with bone marrow stromal cells for the repair of critical bone defects: an experimental study in mice. J Biomed Mater Res A 2009; 90: 1218–1229.
Bujan J, Pascual G, Corrales C et al, Muscle-derived stem cells used to treat skin defects prevent wound contraction and expedite reepithelialization. Wound Repair Regen 2006; 14: 216–223.
Hou L, Wang X, Zhou Y et al, Inhibitory effect and mechanism of mesenchymal stem cells on liver cancer cells. Tumour Biol 2014; 35: 1239–1250.
Koyanagi-Aoi M, Ohnuki M, Takahashi K et al, Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci USA 2013; 110: 20569–20574.
Rodriguez R, Rosu-Myles M, Aráuzo-Bravo M et al, Human bone marrow stromal cells lose immunosuppressive and anti-inflammatory properties upon oncogenic transformation. Stem Cell Rep 2014; 3: 606–619.
Bai S, Kopan R, Zou W et al, NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem 2008; 283: 6509–6518.
Pradel W, Mai R, Gedrange T et al, Cell passage and composition of culture medium effects proliferation and differentiation of human osteoblast-like cells from facial bone. J Physiol Pharmacol 2008; 59 (Suppl 5): 47–58.
Tarnowski M, Koryciak-Komarska H, Czekaj P et al, The comparison of multipotential for differentiation of progenitor mesenchymal-like stem cells obtained from livers of young and old rats. Folia Histochem Cytobiol 2007; 45: 245–254.
McElhinny AS, Li JL, Wu L . Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene 2008; 27: 5138–5147.
Engin F, Lee B . NOTCHing the bone: insights into multi-functionality. Bone 2010; 46: 274–280.
Lin GL, Hankenson KD . Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J Cell Biochem 2011; 112: 3491–3501.
Tao J, Chen S, Lee B . Alteration of Notch signaling in skeletal development and disease. Ann NY Acad Sci 2010; 1192: 257–268.
Ferrari-Toninelli G, Bonini SA, Uberti D et al, Notch activation induces neurite remodeling and functional modifications in SH-SY5Y neuronal cells. Dev Neurobiol 2009; 69: 378–391.
Shutter JR, Scully S, Fan W et al, Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev 2000; 14: 1313–1318.
Gridley T . NOTCH signaling and inherited disease syndromes. Hum Mol Genet 2003; 12: R9–13.
High FA, Epstein JA . The multifaceted role of NOTCH in cardiac development and disease. Nat Rev Genet 2008; 9: 49–61.
Liu J, Sun K, Bai Y et al, Association of three-gene interaction among MTHFR, ALOX5AP and NOTCH3 with thrombotic stroke: a multicenter case-control study. Hum Genet 2009; 125: 649–656.
Koch U, Radtke F . NOTCH and cancer: a double-edged sword. Cell Mol Life Sci 2007; 64: 2746–2762(Erratum in: Cell Mol Life Sci 2008;65:1005).
Fortini ME . Notch signaling: the core pathway and its posttranslational regulation. Dev Cell 2009; 16: 633–647.
Kovall RA . More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 2008; 27: 5099–5109.
Krejcí A, Bernard F, Housden BE et al, Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci Signal 2009; 2: ra1 (Erratum in: Sci Signal 2009;2:er3).
Byth HA, Mchunu BI, Dubery IA et al, Assessment of a simple, non-toxic Alamar blue cell survival assay to monitor tomato cell viability. Phytochem Anal 2001; 12: 340–346.
Mondal R, Koev G, Pilot-Matias T et al, Development of a cell-based assay for high-throughput screening of inhibitors against HCV genotypes 1a and 1b in a single well. Antiviral Res 2009; 82: 82–88.
Sheskin DJ, Handbook of Parametric and Nonparametric Statistical Procedures, 3rd edn. Chapman & Hall/CRC: Boca Raton, 2003, pp 1231.
Millar N, Merlin—Statistics Add-In for Biology Students. Version 2.5. Available at: www.heckgrammar.kirklees.sch.uk(2006).
Ning H, Lin G, Lue TF et al, Neuron-like differentiation of adipose tissue-derived stromal cells and vascular smooth muscle cells. Differentiation 2006; 74: 510–518.
Hoffman RM . The potential of nestin-expressing hair follicle stem cells in regenerative medicine. Expert Opin Biol Ther 2007; 7: 289–291.
da Silva Meirelles L, Chagastelles PC, Nardi NB . Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci 2006; 119: 2204–2213.
Steelman LS, Bertrand FE, McCubrey JA . The complexity of PTEN: mutation, marker and potential target for therapeutic intervention. Expert Opin Ther Targets 2004; 8: 537–550.
Parsons R, Simpson L . PTEN and cancer. Methods Mol Biol 2003; 222: 147–166.
Eng C . PTEN: one gene, many syndromes. Hum Mutat 2003; 22: 183–198.
Chappell WH, Green TD, Spengeman JD et al, Increased protein expression of the PTEN tumor suppressor in the presence of constitutively active Notch-1. Cell Cycle 2005; 4: 1389–1395.
Whelan JT, Forbes SL, Bertrand FE . CBF-1 (RBP-J kappa) binds to the PTEN promoter and regulates PTEN gene expression. Cell Cycle 2007; 1: 80–84.
Waite KA, Eng C . BMP2 exposure results in decreased PTEN protein degradation and increased PTEN levels. Hum Mol Genet 2003; 15: 679–684.
Gruendler C, Lin Y, Farley J et al, Proteasomal degradation of Smad1 induced by bone morphogenetic proteins. J Biol Chem 2001; 276: 46533–46543.
Honda R, Tanaka H, Yasuda H . Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997; 420: 25–27.
Kurose K, Gilley K, Matsumoto S et al, Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet 2002; 32: 355–357.
Stambolic V, MacPherson D, Sas D et al, Regulation of PTEN transcription by p53. Mol Cell 2001; 8: 317–325.
Mayo LD, Dixon JE, Durden DL et al, PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J Biol Chem 2002; 277: 5484–5489.
Tang Y, Eng C . PTEN autoregulates its expression by stabilization of p53 in a phosphatase-independent manner. Cancer Res 2006; 66: 736–742.
Purow BW, Sundaresan TK, Burdick MJ et al, Notch-1 regulates transcription of the epidermal growth factor receptor through p53. Carcinogenesis 2008; 29: 918–925.
Yugawa T, Handa K, Narisawa-Saito M et al, Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol 2007; 27: 3732–3742.
Acknowledgements
The authors acknowledge Dr Anna Sitkowska for technical assistance with microarray assays and Ms Aleksandra Bryzek for technical assistance with some animal work. This study was partially funded by the Institutional funds NN-1-045/07, KNW-1-007/08, and KNW-2-008/08 (to ALS) and NN-1-067/07 (to PC). Some of the equipments used in this work were purchased using EU funds (European Regional Development Fund) within the Sectoral Operational Program ‘Increase of Economic Competitiveness.’ No. WKP 1/1.4/3/2/2005/103/223/565/ 2007/U, Silesian Biopharma Center for Biotechnology, Bioengineering and Bioinformatics, Project no. POIG.02.01.00-00-166/08, and the Operational Program Innovative Economy for 2007–2013, Priority Axis 2. R&D Infrastructure.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Expression, distribution and activation patterns of Notch 1, 2, and Delta1 vary as early as 24-hours following induction of osteogenic conditions. The transcription patterns of several genes known to be linked to osteogenesis, such as Bmps and Egfr, as well as genes regulating the cell cycle and apoptosis follow the Notch pathway activation.
Supplementary information
Rights and permissions
About this article
Cite this article
Urbanek, K., Lesiak, M., Krakowian, D. et al. Notch signaling pathway and gene expression profiles during early in vitro differentiation of liver-derived mesenchymal stromal cells to osteoblasts. Lab Invest 97, 1225–1234 (2017). https://doi.org/10.1038/labinvest.2017.60
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2017.60
This article is cited by
-
Optogenetic control of NOTCH1 signaling
Cell Communication and Signaling (2022)
-
Osteoporosis pathogenesis and treatment: existing and emerging avenues
Cellular & Molecular Biology Letters (2022)
-
Notch signaling in malignant gliomas: supporting tumor growth and the vascular environment
Cancer and Metastasis Reviews (2022)
-
Dose-dependent mechanism of Notch action in promoting osteogenic differentiation of mesenchymal stem cells
Cell and Tissue Research (2020)
-
Contextual Regulation of Skeletal Physiology by Notch Signaling
Current Osteoporosis Reports (2019)