Abstract
Age-related macular degeneration (AMD) is a leading cause of visual impairment and irreversible blindness in most developed countries, affecting about 50 million elderly people worldwide. Retinal pigment epithelial (RPE) cell degeneration is the pathophysiological cause of AMD, leading to geographic atrophy and choroidal neovascularization. We and others have previously identified several polymorphisms on chromosome 10q26 (HTRA1 rs11200638 as well as LOC387715 rs10490924 and c.372_815del443ins54) associated with AMD. In this study, we confirmed the association of our previously identified HTRA1 insertion–deletion (indel) variant (c.34delCinsTCCT) in 195 exudative AMD patients and 390 controls from the Hong Kong Chinese cohort with additional 168 patients and 210 controls from the Chengdu Chinese cohort and followed by studying its biological functions in RPE cells. Genetic analysis verified the higher prevalence of c.34delCinsTCCT allele in control subjects (8.0%) than in AMD patients (1.9%; P=7.87 × 10−5, odds ratio=0.229). This protective effect was validated as the haplotype of the c.34delCinsTCCT allele existed independent of the risk haplotype (P=1.17 × 10−5). In vitro studies showed that recombinant HTRA1 c.34delCinsTCCT variant protein was more localized in the endoplasmic reticulum of RPE cells compared with the wild-type protein, and its secretion was delayed. Moreover, ARPE-19 cells expressing HTRA1 c.34delCinsTCCT variant had higher cell viability, lower cell apoptosis and were less responsive to anoikis, supporting its protective role. We revealed a protective AMD-associated HTRA1 variant in Chinese populations and the biological role of HTRA1 in RPE cell degeneration, indicating its involvement in AMD pathogenesis.
Similar content being viewed by others
Main
Age-related macular degeneration (AMD) is a leading cause of visual impairment and irreversible blindness in most developed countries, affecting about 50 million elderly people worldwide.1 Excessive drusen, the clinical hallmark of AMD, can lead to retinal pigment epithelium (RPE) damage, which instigates geographic atrophy (non-exudative type), vascular endothelial growth factor (VEGF) expression and choroidal neovascularization (CNV; exudative type) with chronic aberrant inflammation.2 AMD is a complex disease influenced by multiple genes and environmental factors. We previously identified a variant (rs11200638) in the promoter region of the high temperature requirement factor A1 (HTRA1) gene on chromosome 10q26 associated with exudative AMD.3, 4 HTRA1 is in strong linkage disequilibrium with an adjacent hypothetical gene LOC387715 (ARMS2), which has also been proposed to be an AMD candidate gene.5 A non-synonymous variant (p.(Ala69Ser); rs10490924:G>T) was suggested to affect the presumptive function of the LOC387715 protein in mitochondria or cytoskeleton.6, 7 In the LOC387715 gene, an insertion–deletion (indel) variant (c.372_815del443ins54), in the same haplotype with the rs11200638 risk allele, downregulates LOC387715 mRNA although it might not correlate with LOC387715 level in the retina or blood.8, 9 Concurrently, this indel allele might also upregulate HTRA1 expression.10 However, a nonsense variant (p.(Arg38Ter); rs2736911:C>T), located in a non-risk haplotype, also led to reduction in LOC387715 mRNA.8, 10 These findings suggested that LOC387715 insufficiency alone may not lead to the development of AMD on 10q26.8
Human HTRA1 belongs to the evolutionarily conserved HtrA family of chymotrypsin-like serine protease with temperature-dependent proteolytic and molecular chaperone activities.11 It was first identified in SV40-transformed fibroblasts and osteoarthritic cartilage12, 13 and predicted to possess a N-terminal secretory signal peptide, a mac25-like domain and a C-terminal HtrA (proteolytic and PDZ) domain. Mutations in the HTRA1 gene are associated with cerebral autosomal-recessive arteriopathy with subcortical infarcts and leukoencephalopathy.14 Downregulation of HTRA1 has been detected in cancers, whereas HTRA1 overexpression enhanced cell apoptosis and reduced cell proliferation in cancer cell lines.15 Upregulation of HTRA1 was reported in placentation,16 preeclampsia,17 arthritis,13 Alzheimer’s disease,18 Duchenne muscular dystrophy19 and idiopathic dilated cardiomyopathy.20 Analogously, HTRA1 mRNA and protein expression are elevated in the lymphocytes and RPE of AMD patients carrying the risk-associated variant,4 while different reported studies had been inconsistent.8 HTRA1 protein expression is elevated in drusen, abnormal RPE and CNV lesion of AMD eyes.4, 21 In addition, we found an association of HTRA1 with VEGF in human vitreous humors and fetal RPE cells upon cell stress and inflammation.22 Collectively, these evidences indicate that HTRA1 could be involved in AMD pathogenesis.
Our previous sequencing study, which involved 163 exudative AMD patients and 183 control subjects, identified an indel variant (c.34delCinsTCCT) in the coding region of HTRA1 gene.23 In the present study, genotyping was extended to increased number of AMD and control subjects in the Hong Kong Chinese cohort. An additional independent Chinese cohort (Chengdu) was also included to validate the association. The properties of the recombinant HTRA1 variant protein were investigated. We aimed to further verify the protective role of this HTRA1 indel variant in AMD.
Materials and methods
Study Subjects
Unrelated Chinese exudative AMD patients and age- and gender-matched control subjects (Supplementary Table S1) were recruited at the Prince of Wales Hospital and the Hong Kong Eye Hospital in Hong Kong (195 patients and 390 controls) and at the Sichuan Provincial People’s Hospital in Chengdu (168 patients and 210 controls). All study subjects were given complete ophthalmoscopic examinations and graded by an international classification and grading system.24 Patients with exudative AMD had non-drusenoid RPE detachment, CNV, serous or hemorrhagic retinal detachments, subretinal or sub-RPE hemorrhage or fibrosis. The control subjects did not have family history of AMD, sign of AMD or any other major eye diseases except mild senile cataracts and low myopia. Their fundi were normal with no drusen, no abnormal RPE change and no foveal reflex. The study protocol was approved by the Ethics Committee for Human Research at the Chinese University of Hong Kong and the Sichuan Provincial People’s Hospital. It was in accordance with the tenets of the Declaration of Helsinki. Informed consent was obtained from the study subjects after explanation of the nature and possible consequences of the study.
HTRA1 Genotyping
Genomic DNA from peripheral venous blood samples of the study subjects was extracted (QIAamp DNA Kit; Qiagen, Germany) according to the supplier’s instructions. The genotype of c.34delCinsTCCT in HTRA1 (ENSG00000166033; Ensembl Genome Browser: http://www.ensembl.org/) was determined by PCR (forward primer: 5′-GTCCCCAAGGCGGCTCGT-3′, reverse primer: 5′-CGCGAAGCTCGGTTCCGAGG-3′)23 and direct sequencing (BigDye Terminator Cycle Sequencing Reaction Kit v3.1; Applied Biosystems, Foster City, CA, USA) on a DNA sequencer (ABI 3130XL; Applied Biosystems).
Meta-Analysis
The meta-analysis of the Hong Kong and Chengdu cohorts was performed by the Review Manager software (RevMan, version 5.2, The Cochrane Collaboration, Copenhagen, Denmark) to generate the Z-scores. The exact P-values with the Z-scores were then calculated using R (v3.0.0, http://cran.r-project.org/). The I2 statistic was used to assess the heterogeneity among studies, which corresponds to no (<25%), low (25%–50%), moderate (50%–75%) and high heterogeneity (≥75%). The random effects model was adopted in the meta-analysis. Summary P<0.05 was considered statistically significant.
Haplotype Analysis
The genotypes of rs2736911 and rs10490924 in LOC387715 and rs11200638 in HTRA1 were collected from previous studies for haplotype construction and estimation of four LOC387715/HTRA1 polymorphisms (http://www.broad.mit.edu/mpg/haploview/, Haploview version 4.2, Massachusetts Institute of Technology, Cambridge, MA, USA).23, 25 Haplotype conditional analysis was performed using PLINK (v1.07; http://pngu.mgh.harvard.edu/purcell/plink/) to test whether the 34delCinsTCCT haplotype had an independent haplotype effect. Moreover, the haplotypes containing rs11200638 and c.34delCinsTCCT of HTRA1 were also determined using allele-specific PCR followed by direct sequencing. DNA from carriers of the heterozygous 34delCinsTCCT genotype, one exudative AMD patient and four control subjects, was analyzed with specific primers (forward primer for rs11200638 G allele: 5′-GA CGCTGCCTTCGTCCG-3′; forward primer for rs11200638 A allele: 5′-GGACGCTGCCTT CGTCCA-3′; and a common reverse primer: 5′-TCGCAGCGGTCTGGGCA-3′).
Bioinformatic Analysis for HtrA1 Signal Peptide
The signal peptide sequences of human HTRA1 wild-type (GenBank accession number: NP_002766.1) and variant (c.34delCinsTCCT) were determined by a prediction program, SOSUIsignal (http://bp.nuap.nagoya-u.ac.jp/sosui/sosuisignal/).26 HTRA1 signal peptide sequences from different species and human HTRA3 signal peptide were aligned by ClustalW (http://www.ebi.ac.uk/clustalw/). The hydrophobicity profile of wild-type and variant signal peptides was predicted by the Kyte–Doolittle algorithm (Protscale tool, http://ca.expasy.org/tools/protscale.html) with a window size of 7.27 The probability score for the central hydrophobic core region of the wild-type and variant signal peptides was estimated by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
Cloning of Recombinant Human HTRA1
A full-length open-reading frame of the HTRA1 gene (NM_002775.4; 1440 base pairs) was cloned into an empty pcDNA6/myc-His A vector (Invitrogen, Carlsbad, CA, USA) between the BamHI and NotI sites (pHis/myc-HTRA1) using specific primers (forward: 5′-TAATGGATCCCCATGCAGATCCCGCGC-3′; reversed: 5′-TAATGCGGCCGCGGGTCAATTTCTTCGGG-3′). Expression construct for the recombinant human HtrA1 variant (pHis/myc-HTRA1-12insSer) was generated by site-directed mutagenesis (QuikChange Lightning Multi Site-Directed Mutagenesis Kit; Strategene, La Jolla, CA, USA) using specific primers (sense: 5′-GCGCCGCTCTTCTCCCGCTGTCCTTGCTGCTGCTGCTGGCGGCG-3′; anti-sense: 5′-CGCCGCCAGCAGCAGCAGCAAGGACAGCGGGAGAAGAGCGGCGC-3′) based on the expression construct for the recombinant human wild-type HtrA1 (pHis/myc-HTRA1). Sequences of both constructs were verified by direct sequencing.
RPE Cell Culture
The human ARPE-19 cell line (CRL-2302, American Type Culture Collection) and primary human fetal RPE cells were maintained in Dulbecco’s modified Eagle’s medium and F-12 nutrient mixture (Gibco BRL, Rockville, MD, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco BRL) and 1 × penicillin/streptomycin (Gibco BRL) at 37 °C in 5% CO2.22 ARPE-19 cells at 90% confluence were transfected with 4 μg HtrA1 expression constructs in 10 μl Lipofectamine-2000 (Invitrogen) and incubated for 12–48 h before further analysis. ARPE-19 cells transfected with empty pcDNA6/myc-His A vector (Invitrogen) were used as control.
HtrA1 Expression Analysis in ARPE-19 Cells
Each of the following experiments was performed in triplicates. Total RNA (RNeasy Extraction Kit; Qiagen) was reverse-transcribed by SuperScript III reverse transcriptase (Invitrogen). HTRA1 mRNA expression was confirmed by PCR.22 HTRA1 protein expression was determined from whole-cell lysate by immunoblotting using mouse monoclonal antibodies against HTRA1 (R&D Systems Inc., Minneapolis, MN, USA). Secreted HTRA1 protein in the serum-free medium collected at specific time points after transfection (12, 18, 24 and 36 h) was concentrated by immunoprecipitation (IP) using mouse monoclonal antibodies against polyhistidine tag (Upstate Biotechnology Inc., Lake Placid, NY, USA) and detected by immunoblotting using mouse monoclonal antibodies against HTRA1 (R&D Systems). HTRA1 in the cell lysate of the HtrA1-transfected ARPE-19 cells was determined to estimate the HtrA1 protein synthetic rate for each construct.
Proteolytic activities of secreted HTRA1 protein were determined by casein digestion.13 Briefly, equal amount of total protein (10 μg) in the medium from HTRA1-transfected ARPE-19 cells were incubated with 5 μg of β-casein (Sigma-Aldrich) in 50 mM Tris/HCl (pH 7.5; Sigma-Aldrich) and 150 mM NaCl (Sigma-Aldrich) at 37 °C. The mixtures, collected at time points 0, 10, 20, 30, 40 and 50 min, were resolved by 15% SDS-PAGE and stained with Coomassie Blue. The protein bands were imaged and the densities were analyzed by the Quantity One Image Analysis software (Bio-Rad). The sample containing buffer and β-casein only was used as the negative control.
Subcellular localization of the recombinant HTRA1 protein was determined by double immunofluorescent labeling. After fixation (3% paraformaldehyde, Sigma-Aldrich), permeation and blocking, the HTRA1-transfected cells were labeled with primary rabbit polyclonal antibodies against giantin (Golgi marker; Covance, Emeryville, CA, USA) or Bip (endoplasmic reticulum (ER) marker; BD Biosciences, San Jose, CA, USA) and secondary antibodies against rabbit IgG conjugated with Alexa Fluor488 (Invitrogen). The cells were further labeled with primary mouse monoclonal antibodies against HTRA1 (R&D Systems) and secondary antibodies against mouse IgG conjugated with Rhodamine Red-X (Invitrogen). The fluorescence signals were visualized under a confocal microscope (TCS SP5; Leica, Germany). For each experiment, 9 fields were captured, and around 200 cells were counted in each field. Percentage of ER localization was calculated by the number of transfected cells with partial overlap signal of HtrA1 with giantin or Bip over the total number of HtrA1-transfected cells.
Cell Survival Analysis
Viability of the HTRA1-transfected ARPE-19 cells at 24 h after transfection was assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (Invitrogen). Absorbance at 570 nm with a reference of 650 nm was measured by a spectrophotometer (Powerwave XS, Bio-Tek Instruments). The percentage of cell viability was determined as OD570 sample/OD570 control × 100%. Cell apoptosis was analyzed by counting the number of transfected ARPE-19 cells with normal and fragmented nuclei (the end stage of apoptosis). HTRA1-transfected cells were identified by immunofluorescent labeling of monoclonal antibodies against HtrA1 (R&D Systems). For each experiment, 9 fields were captured, and around 200 cells were counted in each field. The apoptosis rate was calculated as the number of transfected cells with fragmented nuclei over the total number of HTRA1-transfected cells. Anoikis was induced as described.28 Briefly, equal number of transfected ARPE-19 cells (2 × 105 cells) was transferred to poly-hema-coated plates for 8 h. Cells were allowed to reattach in adhesive plates overnight, followed by Coomassie Blue staining. We previously showed that HTRA1 was upregulated in primary human fetal RPE cells treated with tunicamycin and dithiothreitol (DTT) but downregulated with Z-Leu-Leu-Leu-H (MG132).22 The viability of ARPE-19 cells treated with these cell stress inducers, tunicamycin (5 and 10 μg/ml; Sigma-Aldrich), DTT (1 and 2 mM; Sigma-Aldrich) and MG132 (10 and 20 μM; Sigma-Aldrich), in serum-free medium for 18 h was examined by MTT assay.
Statistical Analysis
Genotypes of the HTRA1 variant between patients and control subjects were compared by the χ2 test. The risk analysis was estimated by odds ratios (OR). Independent T-test was used to compare the mean in the biological studies. All statistical analyses were performed by commercially available software (SPSS, version 16.0; SPSS Inc., Chicago, IL, USA). Significance was defined as P<0.05.
Results
Genotypic Distribution of HTRA1 c.34delCinsTCCT Variant in Two Chinese Cohorts
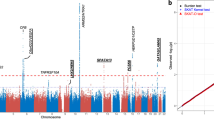
The frequencies of HTRA1 c.34delCinsTCCT indel allele in control subjects were higher than that of exudative AMD patients in the two study cohorts (Hong Kong: 8.0% versus 1.0%, and Chengdu: 8.1% versus 3.0%, respectively). Under the dominant model, the association was significant with exudative AMD in both Hong Kong (P=6.23 × 10−4; OR=0.120, 95% confident interval (CI): 0.028–0.507) and Chengdu cohorts (P=0.035; OR=0.348, 95% CI: 0.126–0.965; Table 1) and with the cohorts combined (P=8.33 × 10−5; OR=0.226, 95% CI: 0.101–0.505). Similarly, under the additive model, the association was also significant in both Hong Kong (P=5.69 × 10−4; OR=0.120, 95% CI: 0.029–0.505) and Chengdu cohorts (P=0.037; OR=0.358, 95% CI: 0.131–0.981) and with the cohorts combined (P=7.87 × 10−5; OR=0.229, 95% CI: 0.103–0.508). Moreover, meta-analysis of the dominant model on our two cohorts (heterogeneity P=0.22) confirmed this significant association (summary P=2.00 × 10−4; OR=0.22, 95% CI: 0.09–0.49; Figure 1). These results indicated that the HTRA1 c.34delCinsTCCT indel variant is protective.
Meta-analysis of HTRA1 34delCinsTCCT variant in Hong Kong and Chengdu Chinese exudative AMD cohorts. The meta-analysis was performed under dominant model. Random effects model was used. Squares indicate study-specific OR. The size of the box is proportional to the sample size of the study. Horizontal lines indicate 95% CI. Diamond indicates summary OR with its corresponding 95% CI.
Haplotype Analysis of LOC387715/HTRA1 Variants
Haplotype analysis of the combined cohorts showed that there were four computed haplotypes constructed for ARMS2 rs2736911 and rs10490924 as well as for HTRA1 rs11200638 and c.34delCinsTCCT. The major haplotypes were the risk (H1) and the wild-type (H2) haplotypes (Table 2). The risk alleles of rs10490924 (T) and rs11200638 (A) were present only in the risk haplotype (H1). The c.34delCinsTCCT indel allele (I), joining with the wild-type alleles of other variants, existed as an independent haplotype (H4). The H4 haplotype (OR=0.095, 95% CI: 0.023–0.399; P=6.23 × 10−5) conferred stronger protection than the wild-type H2 haplotype (OR=0.236, 95% CI: 0.162–0.344; P=2.15 × 10−15). Moreover, the haplotype conditional analysis showed that the haplotype containing c.34delCinsTCCT indel allele exists independent of the risk haplotype (OR=0.097, 95% CI: 0.033–0.285; P=1.17 × 10−5). In addition, the estimated haplotype (H4) was confirmed by the allele-specific PCR of c.34delCinsTCCT and rs11200638. The indel allele of c.34delCinsTCCT was linked to the wild-type allele (G) of rs11200638 but not to the risk allele (A) (Figure 2).
Haplotype analysis of rs11200638 and 34delCinsTCCT. DNA from study subjects with a genotype of heterozygous 34delCinsTCCT and rs11200638 was amplified by PCR using rs11200638 allele-specific primers for direct sequencing. (a) The risk A allele of rs11200638 shared the same haplotype with wild-type C allele of 34delCinsTCCT. (b) The wild-type G allele of rs11200638 shared the same haplotype with indel allele of 34delCinsTCCT. The 34delCinsTCCT indel allele is located in a different haplotype with the risk A allele of rs11200638.
Bioinformatic Analysis for the HTRA1 Signal Peptide
The wild-type C allele of c.34delCinsTCCT coded for a leucine (CUG) at residue 12, while the variant delCinsTCCT allele inserted a serine (UCC) without affecting the leucine (UUG) at residue 13 (Supplementary Figure S1). Based on SOSUIsignal prediction,26 residue 12 lies within the signal peptide of HTRA1 protein. The leucine residue at position 12 is highly conserved among different species (Figure 3a). It resides in the central hydrophobic core region (Figure 3b), which is an indispensable part of the signal peptide.29 According to the Kyte–Doolittle scale,27 hydrophobicity of leucine is 3.80 and serine is −0.80. Insertion of the serine residue in the indel variant was thus predicted to reduce the hydrophobicity (Figure 3b) and the probability score for the hydrophobic region (Figure 3c). It may affect, but not abolish, the property of the HTRA1 signal peptide.
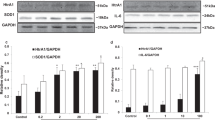
Bioinformatic analysis, expression, proteolytic activity, secretion and ER localization of HTRA1 in ARPE-19 cells. (a) In the alignment of HTRA1 signal peptide sequences from different species, leucine at position 12 was highly conserved. (b) The hydrophobicity profiles of wild-type and variant signal peptides were predicted by the Kyte–Doolittle algorithm with the window size of 7. Signal peptide of HTRA1 variant (dotted line) had lower hydrophobicity than that of wild-type HTRA1 (solid line). (c) Probability scores for the central hydrophobic core regions of the signal peptides were estimated by SignalP 3.0. The HTRA1 variant signal peptide (dotted line) had a less hydrophobic central core region than the wild-type signal peptide (solid line). (d) HTRA1 gene expression for HTRA1-transfected ARPE-19 cells was analyzed by RT-PCR. Comparable band signals were observed in cells transfected with HTRA1. (e) HTRA1 protein in the lysate of HTRA1-transfected ARPE-19 cell was detected by immunoblotting. Comparable band signals were observed. (f) Equal amount of total protein (10 μg) in the serum-free medium from HTRA1-transfected ARPE-19 cells was incubated with 5 μg of β-casein in a buffer of 50 mM Tris/HCl (pH 7.5) and 150 mM NaCl at 37 °C. The reaction mixture at different time points was resolved by 15% SDS-PAGE and visualized by Coomassie Blue. The sample containing only β-casein and buffer was used as a negative control. Secreted HTRA1 protein could digest β-casein, as shown by the time-dependent reduction of uncleaved β-casein and time-dependent accumulation of the cleaved product. In the condition medium of the empty vector-transfected ARPE-19 cells, no cleaved product of β-casein was observed. The accumulation rate of cleaved product of the wild-type HTRA1 protein (solid line) was not differentiated from that of the HTRA1 variant protein (dotted line). (g) Serum-free medium from ARPE-19 cells transfected with pHis/myc-HTRA1 and pHis/myc-HTRA1-12insSer was collected at 12, 18, 24 and 36 h after transfection. Secreted full-length HTRA1 from the medium was concentrated by immunoprecipitation (IP) using mouse anti-His6 antibody and quantified by immunoblotting using mouse monoclonal antibody against HTRA1. Stronger signals of secreted HTRA1 were detected for the ARPE-19 cells transfected with pHis/myc-HTRA1 compared with cells transfected with pHis/myc-HTRA1-12insSer. From the whole-cell lysate, the expression of HTRA1 protein were similar for both constructs. (h) Transfected ARPE-19 cells were fixed and doubly immunolabeled for HTRA1 protein (red fluorescence) and gaintin (marker for Golgi apparatus) or BiP (marker for ER; green fluorescence). Scale bars: 10 μm. About 10% of ARPE-19 cells expressing wild-type HTRA1 showed partial co-localization compared with about 23% of ARPE-19 cells expressing HTRA1 variant. White bar: 24-h posttransfection; black bar: 36-h posttransfection. **P<0.01 compared with wild-type.
Molecular Cloning of the HTRA1 Variant
High HTRA1 mRNA (Figure 3d) and cell lysate HTRA1 protein (Figure 3e) expression levels were detected in the ARPE-19 cells transfected with the HTRA1 wild-type and variant constructs, with no significant difference between the wild type and the variant (data not shown). The HTRA1 protein secreted to the serum-free medium was capable of digesting β-casein, as shown by the time-dependent reduction of uncleaved β-casein and time-dependent accumulation of cleaved product (Figure 3f). In contrast, no cleaved product of β-casein was observed from the condition medium of the empty vector-transfected ARPE-19 cells. The rate of cleaved product accumulation for the wild-type HTRA1 protein could not be differentiated from that for the HTRA1 variant protein, suggesting that the signal peptide variation did not affect its protease activity.
The HTRA1 antibody used in this study detected the full-length HTRA1 protein, which is catalytically active as shown by our β-casein digestion data and a previous study.30 Secreted HTRA1 could not be detected before 12 h of posttransfection (data not shown). From 12 to 36-h posttransfection, stronger signals of secreted HTRA1 were detected for the cells transfected with pHis/myc-HTRA1 compared with that transfected with pHis/myc-HTRA1-12insSer, whereas HTRA1 in the whole-cell lysate was detected similarly for both constructs (Figure 3g). The delayed secretion of HTRA1 variant might be due to the higher percentage of ER localization. The HTRA1 immunofluorescent profiles suggested that HtrA1 protein could be localized mainly in the Golgi apparatus but seldom in ER (Figure 3h). ARPE-19 cells transfected with HTRA1 variant showed more fluorescence signal (2.3-fold, P=0.006) of HTRA1 partially overlapped with that of giantin or Bip at 24 and 36-h posttransfection (22.6% and 23.4%, respectively), compared with those transfected with wild type (9.7% and 10.4%, respectively). The signal peptide property in the HTRA1 variant is altered and thereby affects the trafficking and secretion of the HTRA1 protein.
Cell Survival Analysis of Recombinant Human HTRA1 Protein
ARPE-19 cells expressing wild-type HTRA1 protein had 34.0% reduction of cell viability compared with that transfected with empty vector (P=0.005), while ARPE-19 cells expressing HTRA1 variant protein had 15.4% reduction (P=0.045; Figure 4a). Overexpression of HTRA1 protein therefore reduced viability of the ARPE-19 cells, but higher viability was found in cells expressing HTRA1 variant protein. Moreover, 25.4% of ARPE-19 cells expressed wild-type HTRA1 protein, 15.6% of ARPE-19 cells expressed HTRA1 variant protein and 9.43% of empty vector-transfected ARPE-19 cells possessed fragmented nuclei (Figure 4b). This represented a 2.70-fold increase of apoptotic rate in ARPE-19 cells expressing wild-type HTRA1 protein (P=1.19 × 10−5) and a 1.66-fold increase in ARPE-19 cells expressing HTRA1 variant protein (P=0.001). Ectopic expression of HTRA1 protein would increase cell apoptosis in ARPE-19 cells. Furthermore, ARPE-19 cells expressing HTRA1 variant were less responsive to anoikis when compared with those expressing wild-type protein (Figure 4c). ARPE-19 cells expressing HTRA1 variant could have higher survival rate than those expressing wild-type. We have previously found upregulation of HTRA1 by DTT and tunicamycin but downregulation by MG132.22 Accordingly, we also studied stress-induced human fetal RPE cells to verify the association of enhanced HTRA1 expression with RPE cell viability. The viability of tunicamycin and DTT-treated human fetal RPE cells was reduced (P<0.05), whereas the MG132-treated cells were not affected when compared with the control (Figure 4d).
Cell survival analysis of HTRA1-expressing ARPE-19 cells and stress-induced human fetal RPE cells. (a) Cell viability of HTRA1-expressing ARPE-19 cells was assessed by MTT assay. The percentage of cell viability was expressed as OD570 of ARPE-19 cells expressing wild-type or variant HTRA1/OD570 of ARPE-19 cells transfected with empty vector. ARPE-19 cells expressing HTRA1 variant had higher cell viability than those expressing wild-type HTRA1 (reduction of MTT signal 15.40% versus 34.03%, respectively). (b) A cell undergoing apoptosis was indicated by the fragmented nuclei. HTRA1 protein (red fluorescence) and the DAPI-stained nuclei (blue fluorescence) were labeled by immunocytochemistry. Scale bars: 10 μm. These represented 25.43% in ARPE-19 cells expressing wild-type HTRA1, 15.62% in ARPE-19 cells expressing HTRA1 variant and 9.43% in ARPE-19 cells transfected with empty vector. (c) Anoikis was induced in equal number of transfected ARPE-19 cells and stained by Coomassie Blue. Cells expressing HTRA1 variant had more survival in anoikis than those expressing wild-type. (d) Cell viability of human fetal RPE cells treated with tunicamycin, DTT and MG132 was assessed by MTT assay. Human fetal RPE cells treated with 5 and 10 μg/ml of tunicamycin had 22.6% and 49.7% reduction of MTT signals, respectively, while human fetal RPE cells treated with 1 and 2 mM of DTT had 19.8% and 42.9% reduction of MTT signal, respectively, compared with the control. In contrast, human fetal RPE cells treated with 10 and 20 μM of MG132 had only 6.5% and 3.4% reduction of MTT signal, respectively. *P<0.05, **P<0.01 and ***P<0.001, respectively, compared with control.
Discussion
The HTRA1 c.34delCinsTCCT variant was newly discovered in our previous whole HTRA1 gene sequencing study.23 In this study, we genotyped more AMD and control subjects in our cohort and included additional Chinese cohort from Chengdu in order to verify the frequencies and association patterns of the HTRA1 c.34delCinsTCCT variant in different cohorts. The association of the HTRA1 c.34delCinsTCCT variant in the Chengdu cohort (P=0.035; Table 1) is smaller than that in the Hong Kong cohort (P=6.23 × 10−4), which could be due to smaller number of control subjects used in the Chengdu cohort and few more exudative AMD patients were found in the Chengdu cohort to carry the indel allele. Furthermore, in the haplotype prediction analysis, the TGGC haplotype (H3) is not associated with exudative AMD (P=0.073; Table 2) as the difference in the minor allele frequency of rs2736911 (T) is relatively comparable between the exudative AMD patients (11.5%) and the control subjects (15.9%).
Results from the association study and haplotype analysis indicated a genetic protective effect of the HTRA1 c.34delCinsTCCT variant against AMD (Tables 1 and 2,Figures 1 and 2). The HTRA1 variant was predicted to reduce the characteristic hydrophobicity of the central core region of the signal peptide (Figures 3a–c),29 thereby increasing its ER localization and delaying its secretion (Figures 3g and h). Moreover, ARPE-19 cells expressing the HTRA1 variant had higher cell viability, lower cell apoptosis and less responsive to anoikis compared with those expressing the wild type (Figure 4). Consistent with the genetic results, these biological studies also showed the protective effect of this HTRA1 c.34delCinsTCCT variant.
Enhanced HTRA1 expression attenuates ovarian and trophoblast cell invasion.17, 31 Accordingly, the directly invading choroid capillaries may not be responsible for the HTRA1 upregulation. Moreover, blood vessels had been shown to invade into the cartilaginous condensations and ossification when HTRA1 expression was upregulated,32 indicating that cells with enhanced HTRA1 expression are prone to be invaded by new blood vessels. Therefore, RPE cells should be involved in HTRA1 upregulation in AMD pathogenesis. This postulation could be supported by the elevation of HTRA1 expression in abnormal RPE cells of AMD patients.4, 21
Similar to results in cancer cells,15, 28 overexpression of HTRA1 in ARPE-19 cells in this study reduced cell viability, increased cell apoptosis and responded to anoikis (Figures 4a–c). Moreover, we also found reduced cell viability in human fetal RPE cells treated with DTT and tunicamycin (Figure 4d), in which HTRA1 was upregulated.22 This suggested that HTRA1 induces cell death.33 Accordingly, we propose HTRA1-induced RPE cell death to be a mechanism in AMD pathogenesis as RPE damage is a pathological feature of AMD.2 The report of RPE atrophy in HtrA1 transgenic mice34 further supported our proposition, although HTRA1 could also be involved in regulation of the complement pathway and the amyloid deposition,35 release of cytokine expression36 and ultrastructural changes in the elastic layer of Bruch’s membrane.37
To our knowledge, Golgi localization of recombinant human HTRA1 protein is first reported in this study (Figure 3h) while similar perinuclear staining for HTRA1 has been reported in human cervical cancer CaSki cells.38 There may be a biological role of HTRA1 in Golgi apparatus. Binding of the HTRA1 PDZ domain to cis-Golgi matrix golgin subfamily protein (GM130) has been reported.39 However, subcellular localizations of HTRA1 in different cell types were variable, such as the microtubule-like localization in ovarian cells,31 the nuclear staining for HTRA1 in human cervical cancer CaSki cells, embryonic kidney 293T cells and hepatocyte cells,38 the cytosol location in placental cells16 and the ER localization in HEK293T cells.40 HTRA1 localizations in organelles appear to be cell specific.
In summary, our results, in combination with genetic and in vitro functional studies, revealed a protective HTRA1 variant against AMD. In addition, HTRA1-induced RPE cell death could be a pathogenic mechanism for AMD. Further studies are needed to unwind the biological role of HTRA1 in AMD pathogenesis.
References
Klein R, Peto T, Bird A et al, The epidemiology of age-related macular degeneration. Am J Ophthalmol 2004; 137: 486–495.
de Jong PT . Age-related macular degeneration. N Engl J Med 2006; 355: 1474–1485.
Dewan A, Liu M, Hartman S et al, HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 2006; 314: 989–992.
Yang Z, Camp NJ, Sun H et al, A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science 2006; 314: 992–993.
Fritsche LG, Loenhardt T, Janssen A et al, Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet 2008; 40: 892–896.
Kanda A, Chen W, Othman M et al, A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA 2007; 104: 16227–16232.
Wang G, Spencer KL, Court BL et al, Localization of age-related macular degeneration-associated ARMS2 in cytosol, not mitochondria. Invest Ophthalmol Vis Sci 2009; 50: 3084–3090.
Friedrich U, Myers CA, Fritsche LG et al, Risk- and non-risk-associated variants at the 10q26 AMD locus influence ARMS2 mRNA expression but exclude pathogenic effects due to protein deficiency. Hum Mol Genet 2011; 20: 1387–1399.
Wang G, Spencer KL, Scott WK et al, Analysis of the indel at the ARMS2 3'UTR in age-related macular degeneration. Hum Genet 2010; 127: 595–602.
Yang Z, Tong Z, Chen Y et al, Genetic and functional dissection of HTRA1 and LOC387715 in age-related macular degeneration. PLoS Genet 2010; 6: e1000836.
Spiess C, Beil A, Ehrmann MA . Temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 1999; 97: 339–347.
Zumbrunn J, Trueb B . Primary structure of a putative serine protease specific for IGF-binding proteins. FEBS Lett 1996; 398: 187–192.
Hu SI, Carozza M, Klein M et al, Human HtrA1, an evolutionary conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J Biol Chem 1998; 273: 34406–34412.
Hara K, Shiga A, Fukutake T et al, Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N Engl J Med 2009; 360: 1729–1739.
Baldi A, De Luca A, Morini M et al, The HtrA1 serine protease is down-regulated during human melanoma progression and represses growth of metastatic melanoma cells. Oncogene 2002; 21: 6684–6688.
De Luca A, De Falco M, De Luca L et al, The serine protease HtrA1 is upregulated in the human placenta during pregnancy. J Histochem Cytochem 2004; 52: 885–892.
Ajayi F, Kongoasa N, Gaffey T et al, Elevated expression of serine protease HtrA1 in preeclampsia and its role in trophoblast cell migration and invasion. Am J Obstet Gynecol 2008; 199: 557.e1–557.e10.
Grau S, Baldi A, Bussani R et al, Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc Natl Acad Sci USA 2005; 102: 6021–6026.
Bakay M, Zhao P, Chen J et al, A web-accessible complete transcriptome of normal human and DMD muscle. Neuromuscul Disord 2002; 12: S125–S141.
Colak D, Kaya N, Al-Zahrani J et al, Left ventricular global transcriptional profiling in human end-stage dilated cardiomyopathy. Genomics 2009; 94: 20–31.
Chan CC, Shen D, Zhou M et al, Human HtrA1 in the archived eyes with age-related macular degeneration. Trans Am Ophthalmol Soc 2007; 105: 92–98.
Ng TK, Yam GH, Chen WQ et al, Interactive expressions of HtrA1 and VEGF in human vitreous humors and fetal RPE cells. Invest Ophthalmol Vis Sci 2011; 52: 3706–3712.
Tam PO, Ng TK, Liu DT et al, HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Invest Ophthalmol Vis Sci 2008; 49: 2357–2365.
Bird AC, Bressler NM, Bressler SB et al, An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol 1995; 39: 367–374.
Liang XY, Lai TY, Liu DT et al, Differentiation of exudative age-related macular degeneration and polypoidal choroidal vasculopathy in the ARMS2/HTRA1 locus. Invest Ophthalmol Vis Sci 2012; 53: 3175–3182.
Gomi M, Sonoyama M, Mitaku S . High performance system for signal peptide prediction: SOSUIsignal. Chem-Bio Inform J 2004; 4: 142–147.
Kyte J, Doolittle RF . A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982; 157: 105–132.
He X, Ota T, Liu P et al, Downregulation of HtrA1 promotes resistance to anoikis and peritoneal dissemination of ovarian cancer cells. Cancer Res 2010; 70: 3109–3118.
Nothwehr SF, Gordon JI . Structural features in the NH2-terminal region of a model eukaryotic signal peptide influence the site of its cleavage by signal peptidase. J Biol Chem 1990; 265: 17202–17208.
Eigenbrot C, Ultsch M, Lipari MT et al, Structural and functional analysis of HtrA1 and its subdomains. Structure 2012; 20: 1040–1050.
Chien J, Ota T, Aletti G et al, Serine protease HtrA1 associates with microtubules and inhibits cell migration. Mol Cell Biol 2009; 29: 4177–4187.
Tocharus J, Tsuchiya A, Kajikawa M et al, Developmentally regulated expression of mouse HtrA3 and its role as an inhibitor of TGF-beta signaling. Dev Growth Differ 2004; 46: 257–274.
Zurawa-Janicka D, Skorko-Glonek J, Lipinska B . HtrA proteins as targets in therapy of cancer and other diseases. Expert Opin Ther Targets 2010; 14: 665–679.
Jones A, Kumar S, Zhang N et al, Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc Natl Acad Sci USA 2011; 108: 14578–14583.
An E, Sen S, Park SK et al, Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Invest Ophthalmol Vis Sci 2010; 51: 3379–3386.
Austin BA, Liu B, Li Z et al, Biologically active fibronectin fragments stimulate release of MCP-1 and catabolic cytokines from murine retinal pigment epithelium. Invest Ophthalmol Vis Sci 2009; 50: 2896–2902.
Vierkotten S, Muether PS, Fauser S . Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch's membrane via cleavage of extracellular matrix components. PLoS One 2011; 6: e22959.
Clawson GA, Bui V, Xin P et al, Intracellular localization of the tumor suppressor HtrA1/Prss11 and its association with HPV16 E6 and E7 proteins. J Cell Biochem 2008; 105: 81–88.
Murwantoko Yano M, Ueta Y et al, Binding of proteins to the PDZ domain regulated proteolytic activity of HtrA1 serine protease. Biochem J 2004; 381: 895–904.
Shiga A, Nozaki H, Yokoseki A et al, Cerebral small-vessel disease protein HTRA1 controls the amount of TGF-β1 via cleavage of proTGF-β1. Hum Mol Genet 2011; 20: 1800–1810.
Acknowledgements
We express our greatest appreciation to all the participants in the study. This study was supported by the Health and Medical Research Fund (project number: 12130791 to TKN), the Direct Grant from the Medical Panel, the Chinese University of Hong Kong (grant number: 4054119 to CPP) and the General Research Fund from the Research Grants Council (grant number: 473410 to CPP), Hong Kong and the National Natural Science Foundation of China (grant number: 81025006 to ZY).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
This study describes the genetic and molecular association of a protective HTRA1 insertion-deletion variant (c.34delCinsTCCT) with age-related macular degeneration in Chinese populations. It also reveals that elevated levels of the serine protease HTRA1 induces retinal pigment epithelial cell death, suggesting a potential mechanism for the pathogenesis of age-related macular degeneration.
Supplementary information
Rights and permissions
About this article
Cite this article
Ng, T., Liang, X., Lu, F. et al. Protective effects of an HTRA1 insertion–deletion variant against age-related macular degeneration in the Chinese populations. Lab Invest 97, 43–52 (2017). https://doi.org/10.1038/labinvest.2016.117
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2016.117