Abstract
Pathogenic variants in genes related to channelopathy and cardiomyopathy are the most common cause of sudden unexplained cardiac death. However, few reports have investigated the frequency and/or spectrum of pathogenic variants in these genes in Korean sudden cardiac arrest survivors. This study aimed to investigate the causative genetic variants of cardiac-associated genes in Korean sudden cardiac arrest survivors. We performed exome sequencing followed by filtering and validation of variants in 100 genes related to channelopathy and cardiomyopathy in 19 Korean patients who survived sudden cardiac arrest. Five of the 19 patients (26.3%) had either a pathogenic variant or a likely pathogenic variant in MYBPC3 (n=1), MYH7 (n=1), RYR2 (n=2), or TNNT2 (n=1). All five variants were missense variants that have been reported previously in patients with channelopathies or cardiomyopathies. Furthermore, an additional 12 patients (63.2%) had more than one variant of uncertain significance. In conclusion, pathogenic or likely pathogenic variants in genes related to channelopathy and cardiomyopathy are not uncommon in Korean sudden cardiac arrest survivors and cardiomyopathy-related genes should be included in the molecular diagnosis of sudden cardiac arrest in Korea.
Similar content being viewed by others
Introduction
Sudden cardiac death (SCD) is a major cause of death affecting people of all ages, with an incidence of 1 per 1000 patient-years.1 In the elderly, coronary artery disease is the main cause of SCD, whereas in younger individuals, various other causes are prevalent and a complete postmortem study often fails to determine the cause of death. These autopsy-negative sudden unexplained deaths (SUDs) are frequently explained by heritable cardiac diseases.2 Heritable cardiac channelopathies such as long QT syndrome, catecholaminergic polymorphic ventricular tachycardia and Brugada syndrome (BrS) in a morphologically normal heart affect heart rhythm and cardiac electrical conduction and are associated with sudden cardiac arrest. Additionally, heritable cardiomyopathies, including hypertrophic cardiomyopathy, dilated cardiomyopathy and arrhythmogenic cardiomyopathy, are difficult to diagnose and are often considered indeterminable owing to the uncertain or minimal related changes; therefore, they may account for a significant proportion of SUD cases.3
The identification of dozens of genes related to heart disease coupled with whole-exome sequencing (WES), which enables accurate, thorough and cost-effective genetic analysis of pathogenic variants in the entire library of genes, appears to be useful for comprehensive molecular diagnosis. These advances also have an enormous impact on the surviving relatives of sudden cardiac arrest survivors or victims by enabling preventive treatment or presymptomatic follow-up.4
In a previous study of Korean sudden cardiac arrest survivors with structurally normal hearts, analysis of three cardiac ion channel genes, SCN5A, KCNQ1 and KCNH2, showed that 3 of the 15 patients exhibited a pathogenic variant in SCN5A.5 However, WES and gene panel studies have not yet been used to characterize genes that are related to channelopathy or cardiomyopathy in Korean patients with sudden cardiac arrest. Therefore, in this study, we tried to use WES to identify pathogenic or likely pathogenic variants in 100 genes related to channelopathy or cardiomyopathy in Korean sudden cardiac arrest survivors.
Materials and methods
Study population
Patients who survived sudden cardiac arrest caused by idiopathic ventricular tachycardia or fibrillation were enrolled consecutively from January 2010 to March 2015 at a tertiary care hospital in Seoul, Korea. All sudden cardiac arrest survivor cases were clinically examined by electrocardiogram, two-dimensional echocardiography, coronary angiography or cardiac magnetic resonance imaging. In addition, medical history and family history were recorded. We excluded patients who had abnormal findings in coronary angiography, a positive spasm provocation test with ergonovine (if performed) or a history of coronary artery disease. Written informed consent was obtained from all study subjects. Our institutional review board approved this study (Samsung Medical Center 2014-08-111).
Whole-exome sequencing
Genomic DNA was extracted from peripheral blood leukocytes using the Wizard Genomic DNA Purification Kit following the manufacturer’s instructions (Promega, Madison, WI, USA). SureSelect Human All Exon V5 (Agilent Technologies, Santa Clara, CA, USA) was used for library preparation; sequencing was performed using the Illumina NextSeq500 platform (Illumina Inc., San Diego, CA, USA), generating 2 × 150-bp paired-end reads. Alignment of sequence reads, indexing of the reference genome (hg19) and variant calling with a pipeline was based on Genome Analysis Tool Kit Best Practices. Alignment was performed using BWA-MEM (version 0.7.12); duplicated reads were marked with Picard (version 1.96, http://picard.sourceforge.net); local alignment, base quality recalibration and variant calling were performed with the Genome Analysis Tool Kit, version 3.2–2; and annotation was performed with Variant Effect Predictor, dbNSFP v3.02 (Supplementary Table S1).
Variant filtering steps and data analysis
The number of candidate variants was filtered and prioritized using a four-step strategy to generate a short candidate variant list for experimental validation (Figure 1). Initially, we removed variants with <10 × coverage owing to less reliable variant calls. Next, variants were limited to those with low population frequency, assuming that rare variants are more likely to cause sudden cardiac arrest than common ones. The minor allele frequency (MAF) threshold was carefully chosen based on cardiac disease prevalence (Supplementary Table S2), and variants with an MAF⩾1% in the 1000 Genomes Project database,6 the National Heart, Lung and Blood Institute (NHLBI) Exome Variant Server,7 the Exome Aggregation Consortium (ExAC) database8 or our in-house database of 300 exomes were removed. The third step was to prioritize variants causing nonsynonymous amino-acid changes, start codon alterations, stop loss changes, in-frame insertions/deletions, frameshifts, nonsense variants or changes affecting the consensus splice site sequences. Finally, we performed gene-specific analysis with an in silico gene panel composed of 100 genes including 96 channelopathy and cardiomyopathy genes4 and the recently reported Calmodulin genes (CALM2 and CALM3)9 as the causative genes for long QT syndrome, as well as genes specifically expressed in the heart muscle tissue that are related to cardiomyopathy or channelopathy (NPPA and TNNIK3). These genes were obtained by searching previous publications and the human protein atlas (http://www.proteinatlas.org/, Supplementary Table S3).10
Confirmation and validation of candidate variants
Candidate variants found in WES data were confirmed using standard PCR and Sanger sequencing methods (primer sequences available upon request). Sequence data were aligned to the reference sequence with the Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA).
Criteria for evidence-based classification of candidate variants
Candidate variants were classified according to the standards and guidelines by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP).11 These guidelines recommend for classifying variants into five categories: pathogenic variant (PV), likely pathogenic variant (LPV), variant of uncertain significance (VUS), likely benign variant (BV), and BV based on the combination of many lines of weighted evidences, including population data, computational data, functional data, segregation data, de novo data, allelic data and others. To assess the frequency of a variant in a control or general population, we used the Korean Reference Genome Database,8 which consists of publicly available race-matched control data from whole-genome sequencing of 622 Korean individuals, as well as other public databases such as the 1000 Genomes Project database,6 the NHLBI Exome Variant Server and the ExAC database. A primary literature review was conducted using various sources cited in the Human Gene Mutation Database (HGMD) professional version, release 2016.1 (http://www.hgmd.org/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and PubMed to determine the potential pathogenicity of all identified variants. A variety of in silico tools were used to assess the predicted impact of missense change, including FATHMM (http://fathmm.biocompute.org.uk), SIFT (http://sift.jcvi.org), Polyphen2 (http://genetics.bwh.harvard.edu/pph2), MutationTaster (http://www.mutationtaster.org), MutationAssessor (http://mutationassessor.org), PROVEAN (http://provean.jcvi.org/index.php) and CADD (http://cadd.gs.washington.edu), all of which use missense prediction algorithms, along with GERP (http://mendel.stanford.edu/sidowlab/downloads/gerp/index.html), PhastCons (http://compgen.bscb.cornell.edu/phast/) and PhyloP (http://compgen.bscb.cornell.edu/phast/), all of which use nucleotide-conservation prediction algorithms.
Results
Clinical findings
Twenty Korean patients who survived sudden cardiac arrest were enrolled in the study. One patient was excluded because of positive findings in the spasm provocation test with ergonovine, indicating variant angina. Thus a total of 19 subjects were eligible for analysis. The demographic and clinical characteristics of our cohort are shown in Table 1. There were 16 males (84.2%) and 3 females; the average age at sudden cardiac arrest event was 34.4±15.7 years (range, 8–66 years) with 32.5±16.1 years (range, 8–66 years) for males and 44.7±7.5 years (range, 34–50 years) for females. No patient had a contributing past medical history. One patient had chronic kidney disease (serum creatinine 1.8 mg dl−1). Three had a history of hypertension. No patient had experienced a cardiac event prior to the sudden cardiac arrest. Three patients had a known family history of SCD, and two had a family history of cardiovascular disease such as acute myocardial infarction or congestive heart failure.
WES data analysis and classification of candidate variants
WES was performed in 19 sudden cardiac arrest survivors. The targeted coding sequences of the 100 genes included 89.8 Mb; 89.6% of the target bases were covered by ⩾10 sequence reads. Detailed statistical data are given in Supplementary Table S4.
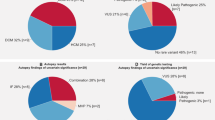
As shown in Table 2, a total of 2 675 336 single-nucleotide variants and insertion/deletions in the exome-targeted region were identified. The number of candidate variants was reduced using a four-step filtration and prioritization strategy to generate a list of candidate variants for further validation. Two patients had no candidate variants, whereas 53 candidate variants were identified in the remaining 17 patients (mean 3.1; range, 1–9). These 53 variants were classified according to the ACMG/AMP guidelines as follows: 2 PVs, 3 LPVs, 34 VUSs and 14 likely BVs. Finally, 2 PVs, 3 LPVs and 34 VUSs were confirmed via Sanger sequencing (Table 2).
The detailed information of candidate variants is listed in Table 3 according to ACMG/AMP guidelines. Five of the 19 patients (26.3%) had either a PV or LPV in MYBPC3 (n=1), MYH7 (n=1), RYR2 (n=2) and TNNT2 (n=1). All five variants were missense variants that were previously reported in patients with channelopathies or cardiomyopathies. The maximum allele frequencies from the ExAC and KRGDB databases for these variants were 0.0003 and 0.004, respectively.12
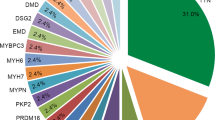
Furthermore, an additional 12 patients (63.2%) had more than one VUS in genes related to channelopathy and cardiomyopathy. A total of 34 VUSs were detected in 15 genes: ANK2, CTNNA3, DSG2, DSP, MYBPC3, PKP2, PRDM16, RBM20, RYR2, SCN2B, SCN5A, TCAP, TNNI3, TNNI3K, and TTN. Among these variants, one in SCN5A was categorized as disease-causing variant for atrial fibrillation in the HGMD while additional five were questionable variants (Table 3).
Discussion
In this study, we identified cardiac disease-associated PV or LPV in 5 of the 19 sudden cardiac arrest survivors via WES focusing on 100 channelopathy- and cardiomyopathy-related genes. In one previous study of 57 families in London with a member who exhibited sudden arrhythmic death syndrome, inherited arrhythmia syndrome was detected in 53% of families via cardiogenetic screening for KCNQ1, KCNE1, KCNH2, KCNE2, SCN5A, ANK215, KCNJ2, CAV3, CASQ2 and RYR2.13 Moreover, in a gene-specific analysis of 117 channelopathy-, cardiomyopathy- or metabolic disorder susceptibility-related genes in 14 consecutively referred Caucasian autopsy-negative SUD in the young victims, 8 ultra-rare variants absent in 3 publically available exome databases were identified in 6 genes in 7 of the 14 cases (50%).3 Furthermore, WES identified potentially lethal cardiac disease-associated variants in three of the five SUD victims in one study conducted at the Zurich Institute of Forensic Medicine in Switzerland.2 In another investigation in the Netherlands, 17 of the 43 (40%) consecutive families with at least one SUD victim who died at ⩽40 years of age had inherited cardiac disease.14 Compared with these data, we observed a smaller proportion of inherited cardiac disease in Korean sudden cardiac arrest survivors and a lower diagnostic yield. We note that our study population consisted of survivors of sudden cardiac arrest, unlike the victims of SCD included in the abovementioned studies. The difference of study subjects might affect the results, although further studies are needed to explore this difference.
Of the five cardiac disease-associated PV or LPV identified in our cohort, two were detected in channelopathy genes and three were detected in cardiomyopathy genes. Several studies investigating cardiac channelopathy-associated genes causing lethal cardiac arrhythmia in SUD cases without any underlying structural pathology revealed that 26–53% of all cases were diagnosed with inherited cardiac channelopathies.13, 15, 16 However, WES studies of cardiomyopathy- and channelopathy-related genes in autopsy-negative SUD cases revealed that 6 of the 14 studied cases harbored rare variants in cardiomyopathy-associated genes; interestingly, only 1 case exhibited a variant in a cardiac channelopathy gene, and 11 cardiomyopathy-related variants and 6 channelopathy-related variants were observed in other cases.3, 17 Autopsy or macroscopic evaluation may fail to identify areas with sufficient abnormal myocardium. Moreover, structurally faulty myocardial conditions genetically induced by cardiomyopathy genes may cause sudden death, even when insufficient to meet diagnostic criteria at a microscopic level.13 In our cohort, two of the three patients (SCAS-1 and SCAS-2) with LPV in cardiomyopathy genes had no evidence of cardiomyopathy when evaluated on two-dimensional echocardiography and the remaining patient diagnosed with hypertrophic cardiomyopathy showed a mild hypertrophic manifestation of the cardiac septum. Therefore, minimal structural abnormalities that are deemed inconclusive or even missed completely may underlie a significant proportion of SUD cases. A study by Maron et al.18 also showed that SCD may be the first manifestation of disease. Therefore, evaluation of cardiomyopathy genes should be considered in patients with a history of life-threatening sudden cardiac arrest and in the relatives of these patients, even when no evidence of cardiomyopathy is present. It is of note that SCAS-2 patient showed echocardiography patterns that were consistent with BrS at he time of sudden cardiac arrest resuscitation but was normalized at follow-up echocardiography test. This patient lacked symptoms that are characteristic to BrS such as palpitation, presyncope and syncope. No variants were identified in BrS-related genes either. Taken together, this patient might have Brugada phenocopy rather than BrS.19
Probands with PV or LPV in cardiomyopathy genes had no family history of cardiac disease except one daughter of SCAS-1 patient who was diagnosed as congestive heart failure. Although periodic review of family history is required, confirming cardiac disease in relatives can be complicated by many factors, including failure to undergo appropriate cardiac screening, reduced penetrance, early death from other causes before the onset of HCM and/or other social issues, and thus it may not be possible to confirm whether a proband with HCM is truly a simplex case or not (https://www.ncbi.nlm.nih.gov/books/NBK1768/). In addition, a PV or LPV in a proband with HCM may occur de novo. According to the study by Morita et al.,20 the proportion of cases caused by de novo PV was unknown but could be estimated to be 30%, which were detected most commonly in MYH7 and MYBPC3. It is also possible that the same PV that is shared by unrelated hypertrophic cardiomyopathy individuals might be due to independent occurrence of de novo variants.21
Perturbation of cardiac ion channels, which are critical in cardiac electrophysiology, can result in SCD owing to fatal ventricular arrhythmias.22 The Heart Rhythm Society/European Heart Rhythm Association guidelines state that ‘in the setting of autopsy-negative SUDs, comprehensive or targeted (RYR2, KCNQ1, KCNH2, and SCN5A) ion channel genetic testing may be considered in an attempt to establish probable cause and manner of death and to facilitate the identification of potentially at-risk relatives and is recommended if circumstantial evidence points toward a clinical diagnosis of long QT syndrome or catecholaminergic polymorphic ventricular tachycardia specifically’.23 Compared with the role of cardiac channelopathy-associated genes in Caucasian SUD cases, KCNQ1 and KCNH2 seem to be very rare causative genes in Korean patients with sudden cardiac arrest.13, 16, 17 Similarly, a previous study of Korean survivors of sudden cardiac arrest revealed no pathogenic variants in KCNQ1 or KCNH2.
Interestingly, MYBPC3-Glu334Lys variant detected in SCAS-2 patient has an MAF of 0.004 in KRGDB. Although this MAF seems to be high compared with estimated disease prevalence, the penetrance of MYBPC3 variants is known to be reduced.24 According to the recently published studies, MAF of some variants that were previously reported to be pathogenic in the HGMD and/or ClinVar might be >1% either globally or in ethnicity-matched controls.25, 26 Although some of this excess can be attributed to BVs falsely assigned as pathogenic, other variants have genuine effects on disease susceptibility but confer reduced lifetime risks or penetrance. Therefore, it is no wonder that a variant with relatively high MAF than expected could be the cause of genetic disease owing to the reduced penetrance.
Most of the filtered variants from the WES data were categorized as VUSs according to the ACMG/AMP guidelines because they lacked sufficient evidences to demonstrate pathogenicity (such as de novo data, segregation data or functional data) as they were novel variants or had not been evaluated further. Although they exhibited very low MAF in the various population databases, their disease associations have not yet been conclusively determined.
Of note, this is the first study that used WES to investigate cardiac-specific genes associated with channelopathy or cardiomyopathy in Korean sudden cardiac arrest survivors. Using WES, we found that 5 of the 19 Korean sudden cardiac arrest survivors had PV or LPV in four genes, MYBPC3, MYH7, RYR2 and TNNT2. Korean sudden cardiac arrest survivors appear to exhibit a smaller proportion of inherited cardiac disease and a lower diagnostic yield compared with subjects in other studies. However, this study has some limitations in that only survivors were investigated. According to 2015 National Emergency Department Information System for Cardiac Arrest data of Korea, 9.6% patients survived to discharge among resuscitation-attempted out-of-hospital cardiac arrests.27 We also conclude that cardiomyopathy may be a significant cause of sudden cardiac arrest even in the absence of clinical findings, because channelopathy- and cardiomyopathy-associated genes were detected at similar proportions. However, further studies with larger Korean cohorts experiencing sudden cardiac arrest are needed to confirm the detection rate of these pathogenic variants and to definitively determine the causative cardiac-associated genes in Korean populations. Nevertheless, the results of this study would be useful for implementing cardiogenetic screening in the general Korean population.
References
Straus, S. M., Bleumink, G. S., Dieleman, J. P., van der Lei, J., Stricker, B. H. & Sturkenboom, M. C. The incidence of sudden cardiac death in the general population. J. Clin. Epidemiol. 57, 98–102 (2004).
Neubauer, J., Haas, C., Bartsch, C., Medeiros-Domingo, A. & Berger, W. Post-mortem whole-exome sequencing (WES) with a focus on cardiac disease-associated genes in five young sudden unexplained death (SUD) cases. Int. J. Legal Med. 130, 1011–1021 (2016).
Narula, N., Tester, D. J., Paulmichl, A., Maleszewski, J. J. & Ackerman, M. J. Post-mortem whole exome sequencing with gene-specific analysis for autopsy-negative sudden unexplained death in the young: a case series. Pediatr. Cardiol. 36, 768–778 (2015).
Saenen, J. B., Van Craenenbroeck, E. M., Proost, D., Marchau, F., Van Laer, L., Vrints, C. J. et al. Genetics of sudden cardiac death in the young. Clin. Genet. 88, 101–113 (2015).
Son, M. K., Ki, C. S., Park, S. J., Huh, J., Kim, J. S. & On, Y. K. Genetic mutation in Korean patients of sudden cardiac arrest as a surrogating marker of idiopathic ventricular arrhythmia. J. Korean Med. Sci. 28, 1021–1026 (2013).
Abecasis, G. R., Auton, A., Brooks, L. D., DePristo, M. A., Durbin, R. M., Handsaker, R. E. et al. An integrated map of genetic variation from 1092 human genomes. Nature 491, 56–65 (2012).
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, USA. http://evs.gs.washington.edu/EVS/. (Accessed 1 May 2016).
Exome Aggregation Consortium (ExAC) Cambridge, MA, USA. http://exac.broadinstitute.org. (Accessed 1 May 2016).
Boczek, N. J., Gomez-Hurtado, N., Ye, D., Calvert, M. L., Tester, D. J., Kryshtal, D. O. et al. Spectrum and prevalence of CALM1-, CALM2-, and CALM3-encoded calmodulin variants in long QT syndrome and functional characterization of a novel long QT syndrome-associated calmodulin missense variant, E141G. Circ. Cardiovasc. Genet. 9, 136–146 (2016).
The Human Protein Atalas (2016). http://www.proteinatlas.org. (Accessed 15 June 2016).
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Korean Reference Genome Database (KRGDB) Cheongju-si, Chungcheongbuk-do, Korea http://152.99.75.168/KRGDB. (Accessed 1 May 2016).
Behr, E. R., Dalageorgou, C., Christiansen, M., Syrris, P., Hughes, S., Tome Esteban, M. T. et al. Sudden arrhythmic death syndrome: familial evaluation identifies inheritable heart disease in the majority of families. Eur. Heart J. 29, 1670–1680 (2008).
Tan, H. L., Hofman, N., van Langen, I. M., van der Wal, A. C. & Wilde, A. A. Sudden unexplained death: heritability and diagnostic yield of cardiological and genetic examination in surviving relatives. Circulation 112, 207–213 (2005).
Stattin, E. L., Westin, I. M., Cederquist, K., Jonasson, J., Jonsson, B. A., Morner, S. et al. Genetic screening in sudden cardiac death in the young can save future lives. Int. J. Legal Med. 130, 59–66 (2016).
Tester, D. J., Medeiros-Domingo, A., Will, M. L., Haglund, C. M. & Ackerman, M. J. Cardiac channel molecular autopsy: insights from 173 consecutive cases of autopsy-negative sudden unexplained death referred for postmortem genetic testing. Mayo Clin. Proc. 87, 524–539 (2012).
Hata, Y., Kinoshita, K., Mizumaki, K., Yamaguchi, Y., Hirono, K., Ichida, F. et al. Postmortem genetic analysis of sudden unexplained death syndrome under 50 years of age: a next-generation sequencing study. Heart Rhythm 13, 1544–1551 (2016).
Maron, B. J., Olivotto, I., Spirito, P., Casey, S. A., Bellone, P., Gohman, T. E. et al. Epidemiology of hypertrophic cardiomyopathy-related death: revisited in a large non-referral-based patient population. Circulation 102, 858–864 (2000).
Gottschalk, B. H., Anselm, D. D., Brugada, J., Brugada, P., Wilde, A. A., Chiale, P. A. et al. Expert cardiologists cannot distinguish between Brugada phenocopy and Brugada syndrome electrocardiogram patterns. Europace 18, 1095–1100 (2016).
Morita, H., Rehm, H. L., Menesses, A., McDonough, B., Roberts, A. E., Kucherlapati, R. et al. Shared genetic causes of cardiac hypertrophy in children and adults. N. Engl. J. Med. 358, 1899–1908 (2008).
Seidman, J. G. & Seidman, C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104, 557–567 (2001).
Albert, C. M., MacRae, C. A., Chasman, D. I., VanDenburgh, M., Buring, J. E., Manson, J. E. et al. Common variants in cardiac ion channel genes are associated with sudden cardiac death. Circ. Arrhythm. Electrophysiol. 3, 222–229 (2010).
Ackerman, M. J., Priori, S. G., Willems, S., Berul, C., Brugada, R., Calkins, H. et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace 13, 1077–1109 (2011).
Page, S. P., Kounas, S., Syrris, P., Christiansen, M., Frank-Hansen, R., Andersen, P. S. et al. Cardiac myosin binding protein-C mutations in families with hypertrophic cardiomyopathy: disease expression in relation to age, gender, and long term outcome. Circ. Cardiovasc. Genet. 5, 156–166 (2012).
Minikel, E. V., Vallabh, S. M., Lek, M., Estrada, K., Samocha, K. E., Sathirapongsasuti, J. F. et al. Quantifying prion disease penetrance using large population control cohorts. Sci. Transl. Med. 8, 322ra329 (2016).
Lek, M., Karczewski, K. J., Minikel, E. V., Samocha, K. E., Banks, E., Fennell, T. et al. Analysis of protein-coding genetic variation in 60706 humans. Nature 536, 285–291 (2016).
Yang, H. J., Kim, G. W., Kim, H., Cho, J. S., Rho, T. H., Yoon, H. D. et al. Epidemiology and outcomes in out-of-hospital cardiac arrest: a report from the NEDIS-based cardiac arrest registry in Korea. J. Korean Med. Sci. 30, 95–103 (2015).
Acknowledgements
This study was supported by Samsung Biomedical Research Institute grants (SMO1161371 and SMX1161381).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of Interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Song, J., Kang, JS., Kim, YE. et al. Identification of pathogenic variants in genes related to channelopathy and cardiomyopathy in Korean sudden cardiac arrest survivors. J Hum Genet 62, 615–620 (2017). https://doi.org/10.1038/jhg.2017.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.8
This article is cited by
-
Exploring TTN variants as genetic insights into cardiomyopathy pathogenesis and potential emerging clues to molecular mechanisms in cardiomyopathies
Scientific Reports (2024)
-
Unexplained cardiac arrest: a tale of conflicting interpretations of KCNQ1 genetic test results
Clinical Research in Cardiology (2018)