Abstract
Mutations in CLDN14, encoding tight junction protein claudin 14, cause profound deafness in mice and humans. We identified a Pakistani family, in which the affected individuals were homozygous for a known pathogenic mutation c.254 T>A resulting in p.V85D substitution in CLDN14; however, in contrast to the previously reported families with mutations in CLDN14, most of the affected individuals in this family exhibit only a severe hearing loss (HL). In order to identify the contribution of CLDN14 to less than profound deafness, we screened for mutations of CLDN14 in 30 multiplex and 57 sporadic cases with moderately severe to severe HL from Pakistan. We identified one other affected individual homozygous for p.V85D substitution. Comparison of audiometric data from all patients indicates that mutations in CLND14 cause varying degrees of HL, which may be enhanced at high frequencies. This suggests that a modifier can reduce the severity of HL associated with mutations of CLDN14. Our data indicate that mutations in CLDN14 should be explored when considering the etiology of less severe HL.
Similar content being viewed by others
Main
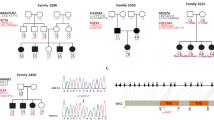
CLDN14 encodes a protein, which participates in the formation of tight junctions in different epithelial cells including those of the cochlear sensory epithelia. Patients with mutations in CLDN14 and Cldn14 knockout mice are profoundly deaf.1, 2 In order to determine the etiology of less severe hearing loss (HL) in Pakistan, we recruited 30 consanguineous families with multiple affected individuals and 57 sporadic cases with moderate to severe HL (50–90 dB HL) with the help of audiologists and schools for special children. After institutional review board approval and written informed consent was acquired, DNA samples were obtained from blood samples of all participants. Linkage analyses for 45 of the known loci of recessively inherited HL were performed for family HLRB5 by genotyping fluorescently labeled microsatellite markers, including D21S1252 and D21S167 for DFNB29. All other families and 57 sporadic cases with HL were similarly checked for linkage or homozygosity for markers flanking CLDN14. Only HLRB5 was consistent with linkage to CLDN14 (Figure 1a), while the single affected individual SA18 was homozygous for the genotyped markers. Sequencing revealed the same mutation c.254 T>A (p.V85D) in CLDN14 segregating in affected individuals in HLRB5 and SA18 (Figure 1b). Affected individuals in family HLRB5 and individual SA18 were homozygous for the same allele linked to the deafness phenotype for marker D21S1252 and also shared single nucleotide polymorphisms in CLDN14 (Table 1), which indicates that this mutation probably arose on a common ancestral chromosome. However, carriers for this mutation are not frequent in the population, as we did not identify this variant in 100 control DNA samples from Sheikhupura (200 chromosomes).
(a) Pedigree with haplotype data for family HLRB5. Solid symbols denote affected individuals. Results of genotyping are shown for two microsatellite markers. The deafness-associated haplotype is shaded in gray. Alleles for each marker are denoted by letters. Allele sizes in base pairs are: D21S1252; A, 249; B, 247; C, 245; D, 239; E, 237; D21S167; A, 162; B, 154; C, 152; D, 146; E, 138; F, 132. (b) Sequence trace files for the c.254 T>A mutation observed in family HLRB5 and SA18 from a normal and affected sample. The mutation is indicated by an arrow in the trace from the affected individual. The normal and mutated codons are underlined in the respective traces.
Recently, inheritance of a synonymous single nucleotide polymorphism in CLDN14 was reported to be associated with kidney stones and low bone mineral density.3 Therefore, we examined three members of family HLRB5 for kidney stones. However, renal ultrasounds failed to detect kidney stones in two affected individuals, V:9 and V:11, who were homozygous for the mutation, and one normal individual, V:7, who was heterozygous for the c.254 T>A mutation.
Pure-tone air conduction averages (PTA) for hearing thresholds at 500, 1000, 2000 and 4000 Hz were calculated to compare the severity of deafness of all affected individuals. HL thresholds were classified as normal (0–25 dB HL), mild (26–40 dB HL), moderate (41–55 dB HL), moderately severe (56–70 dB HL), severe (71–90 dB HL) and profound (>90 dB HL).4 Ten individuals (ages 10–22 years) in family HLRB5 are hearing impaired. According to parents and self-reports, HL was not progressive. The audiometric data for all affected individuals in family HLRB5 gathered 2 years apart suggest that there is no deterioration of hearing. However, HL may progress slowly and additional time is needed to conclusively establish the stability of hearing thresholds.
The best hearing among the affected individuals in family HLRB5 is of a 10-year-old child, V:3, PTA 63 dB HL (moderately severe HL), whereas the worst is of a 15-year-old child, V:13, PTA 91 dB HL (profound deafness) (Figure 2). All other affected individuals in family HLRB5 and SA18 have a severe HL, PTA 74–88 dB HL (Figure 2). The previously reported families with p.V85D mutation have profound deafness (PTA 93–100 dB HL). Thus, the HL associated with mutations in CLDN14 can range from moderately severe to profound deafness.
(a) Audiograms for all affected family members of family HLRB5 and SA18. Age at time of first audiometry is indicated on top of each audiogram. ‘○’ indicates air conduction for right ear, whereas ‘ × ’ indicates air conduction for left ear. (b) Average thresholds at pure tones (500, 1000, 2000 and 4000 Hz) for the better hearing ears in family HLRB5 (circles), individual SA18 (triangle) and previously published families (squares) with mutations in CLDN14. The degree of hearing loss (HL) is different across and within families with mutations in CLDN14. All families have the same mutation, except for family PKSN6.
Variations can also be seen at different frequencies when comparing audiometric data from all families with CLDN14 mutations. At low frequencies (250 and 500 Hz), the HL ranges from moderate to severe. At conversational frequencies (500, 1000 and 2000 Hz) HL ranges from moderately severe to profound, whereas at high frequencies it ranges from severe to profound. However, for family HLRB5, the loss at high frequencies is still less than that reported earlier, with 7 out of 10 individuals responding to these tones, whereas none of the patients in the previously published reports responded to the 8000 Hz frequency.5
This study shows that individuals with mutations of CLDN14 may have different degrees of HL and the loss is greater at high frequencies. Some preservation of low-frequency hearing with a greater degree of high-frequency HL may indicate involvement of CLDN14 in deafness, a finding similar to the HL associated with mutant alleles of GJB2.6 Although CLDN14 is not a contributor to profound deafness in Turkey, Tunisia and Spain,7, 8, 9 it is possible that some cases of less severe HL in these and other countries could be attributed to CLDN14 mutations.
p.V85D is probably a null mutation as functional assays show that its ectopic expression in fibroblast cell lines results in mislocalization of this mutant CLDN14 and its absence from plasma membranes in contrast to the wild-type protein.10 These data suggest that in patients with p.V85D mutation, the dysfunction is due to loss of CLDN14 from tight junctions. It is known that CLDN14 is absent from tight junctions in Cldn14 knockout mice leading to progressive loss of hair cells.2 Mutations in CLDN14 resulting in HL may similarly cause hair cell loss in humans. However, the different degree of HL in humans with CLDN14 mutations indicates that the extent of hair cell loss may vary. In Cldn14−/− mice, variation in hair cell loss is seen with the base of the cochlea affected earlier than the apex. This may also be the case in humans. As the apex of the cochlea responds to low frequency sounds, it may account for the residual hearing at the 4000 and 8000 Hz observed in the individuals reported in this study. Alternately, variations resulting in regulation of another inner ear-expressed claudin may partially compensate for the loss of CLDN14.
Our research indicates that the same pathogenic mutations may not result in identical phenotypic severity and genetic or epigenetic factors may modify the clinical course of this genetic disorder. The search for modifier genes is difficult in humans,11 although a few modifiers for deafness have been mapped or cloned using linkage analyses or candidate gene approaches.12 In family HLRB5, one individual is profoundly deaf and other individuals exhibit moderately severe to severe HLs. A clear demarcation of phenotype is absent, making linkage analyses for mapping a modifier locus difficult. However, mice are more amenable to search for modifiers of different phenotypes and many genes that modify effects of deafness causing genes have been identified.12 Therefore transferring Cldn14 null allele to different genetic backgrounds and isolating the most and least affected strains may help in identification of modifying variants in one or more causative genes.
References
Wilcox, E. R., Burton, Q. L., Naz, S., Riazuddin, S., Smith, T. N., Ploplis, B. et al. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell 104, 165–172 (2001).
Ben-Yosef, T., Belyantseva, I. A., Saunders, T. L., Hughes, E. D., Kawamoto, K., Van Itallie, C. M. et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum. Mol. Genet. 12, 2049–2061 (2003).
Thorleifsson, G., Holm, H., Edvardsson, V., Walters, G. B., Styrkarsdottir, U., Gudbjartsson, D. F. et al. Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat. Gene. 41, 926–930 (2009).
Katz, J. Handbook of Clinical Audiology (Williams & Wilkins, Baltimore, 1994).
Ahmed, Z. M., Riazuddin, S., Friedman, T. B., Riazuddin, S., Wilcox, E. R. & Griffith, A. J. Clinical manifestations of DFNB29 deafness. Adv. Otorhinolaryngol. 61, 156–160 (2002).
Wilcox, S. A., Saunders, K., Osborn, A. H., Arnold, A., Wunderlich, J., Kelly, T. et al. High frequency hearing loss correlated with mutations in the GJB2 gene. Hum. Genet. 106, 399–405 (2000).
Uyguner, O., Emiroglu, M., Uzumcu, A., Hafiz, G., Ghanbari, A., Baserer, N. et al. Frequencies of gap- and tight-junction mutations in Turkish families with autosomal-recessive non-syndromic hearing loss. Clin. Genet. 64, 65–69 (2003).
Arican, S. T., Incesulu, A., Inceoglu, B. & Tekin, M. Alterations in the GJB3 and CLDN14 genes in families with nonsyndromic sensorineural hearing loss. Genet. Couns. 16, 309–311 (2005).
Belguith, H., Tlili, A., Dhouib, H., Ben Rebeh, I., Lahmar, I., Charfeddine, I. et al. Mutation in gap and tight junctions in patients with non-syndromic hearing loss. Biochem. Biophys. Res. Commun. 385, 1–5 (2009).
Wattenhofer, M., Reymond, A., Falciola, V., Charollais, A., Caille, D., Borel, C. et al. Different mechanisms preclude mutant CLDN14 proteins from forming tight junctions in vitro. Hum. Mutat. 25, 543–549 (2005).
Génin, E., Feingold, J. & Clerget-Darpoux, F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum. Genet. 124, 357–368 (2008).
Johnson, K R., Zheng, Q. Y. & Noben-Trauth, K. Strain background effects and genetic modifiers of hearing in mice. Brain Res. 1091, 79–88 (2006).
Acknowledgements
We thank the families for participating in the study. We are grateful to Mr Khalil Hashmi for facilitating sample collection from patients. We thank Professor M Akram Dogar and Professor Adul Lateef Mughal for sample collection from unaffected control samples from Sheikhupura and Dr Andrew Griffith for his expert opinion. We are indebted to Dr Thomas B Friedman for his valuable comments on the paper. This research was supported by Grant number R01TW007608 from the Fogarty International Center and National Institute of Deafness and other Communication Disorders, National Institutes of Health, USA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Bashir, R., Fatima, A. & Naz, S. Mutations in CLDN14 are associated with different hearing thresholds. J Hum Genet 55, 767–770 (2010). https://doi.org/10.1038/jhg.2010.104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.104
Keywords
This article is cited by
-
Molecular genetic landscape of hereditary hearing loss in Pakistan
Human Genetics (2022)
-
A common variant in CLDN14 causes precipitous, prelingual sensorineural hearing loss in multiple families due to founder effect
Human Genetics (2017)
-
Phenotypic variability of CLDN14 mutations causing DFNB29 hearing loss in the Pakistani population
Journal of Human Genetics (2013)
-
The c.42_52del11 Mutation in TPRN and Progressive Hearing Loss in a Family from Pakistan
Biochemical Genetics (2013)