Abstract
Three new β-class milbemycins, 13α-hydroxy-4-ethy1 milbemycin β3 (1), 13α-hydroxy-25-ethy1 milbemycin β3 (2), 13α-hydroxy milbemycin β3 (3), were isolated from the broth of the genetically engineered strains Streptomyces avermitilis MHJ1011, whose aveA1 gene was replaced by milA1 gene seamlessly. Their structures were determined on the basis of extensive spectroscopic analysis and comparison with data from the literature. These three compounds, especially compound 1, exhibited potent acaricidal activity.
Similar content being viewed by others
Introduction
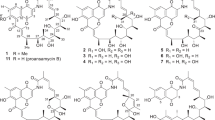
Polyketide synthases have been attractive targets of biosynthetic engineering to make ‘unnatural’ natural products. By modification or deletion one or more of the genes encoding polyketide domains or modules, the genetically engineered strain was constructed1 and exploited to make large numbers of new natural products, such as the analogs of erythromycin,2, 3 amphotericin4, 5 and daptomycin.6 In our previous work, a genetically engineered strain Streptomyces avermitilis MHJ1011, whose aveA1 gene was seamlessly replaced by milA1 gene from Streptomyces hygroscopicus, has been constructed with the aim of obtaining new 16-membered macrolactone compounds. This led to the isolation of tenvermectins A and B (Figure 1) with excellent insecticidal activity.7 The in-depth chemical study of fermentation broth of the strain S. avermitilis MHJ1011 resulted in identification of three new β-class milbemycins (1–3, Figure 1). Herein, we describe the fermentation, isolation, structural elucidation and acaricidal activity of the three new compounds.
Results and discussion
Structural elucidation
In the course of fermentation and isolation of tenvermectins A and B from the strain S. avermitilis MHJ1011, the by-products were pooled and a crude extract was obtained. The pooling crude extract was isolated by silica gel column chromatography and semi-preparative HPLC to afford three new compounds (1–3).
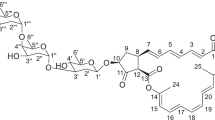
The molecular formula of compound 1 was established to be C32H44O6 as deduced from the HRESI-MS m/z 525.3206 [M+H]+ (calcd for C32H45O6 525.3211) and 13C NMR data (Table 1). The 1H NMR spectrum (Table 1) of 1 displayed three doublet aliphatic methyls at δH 0.84 (3H, d, J=6.5 Hz), 1.14 (3H, d, J=6.2 Hz), 1.21 (3H, d, J=6.9 Hz), two olefinic methyl signals at δH 1.64 (3H, brs) and 2.07 (3H, s), one trans-double bond at δH 5.44 (1H, dd, J=15.1, 9.5 Hz), 6.16 (1H, dd, J=15.1, 10.8 Hz) and two downfield proton signals at δH 7.35 (1H, s) and 6.60 (1H, s). The 13C NMR and HMQC spectra revealed 32 carbon resonances, including an ester carbonyl carbon at δ 169.6 (s), a ketal carbon at δ 97.8 (s), four oxygenated methines at δC 79.1 (d), 71.3 (d), 68.1 (d) and 67.6 (d), two aliphatic methines at δC 40.4 (d) and 36.6 (d), six methyls at δC 19.4 (q), 18.2 (q), 18.1 (q), 17.9 (q), 15.1 (q) and 13.6 (q) in addition to five aliphatic methylenes and 14 sp2 carbons. By detailed comparison of the NMR data of 1 with those of milbemycin β3,8, 9 it was revealed that 1 was similar to milbemycin β3. The differences between 1 and milbemycin β3 were that a hydroxyl group substituted at C-13 and an ethyl group situated at C-4 in 1. The 1H-1H COSY correlation of δH 2.61/ δH 4.02, δH 1.23/δH 2.60, and the observed HMBC correlations from δH 1.21 and 1.64 to δC 79.1, from δH 1.23 to δC 22.5, 128.7 (Figure 2) supported the assignment. Compared with milbemycin β3, the 30 mass unit enhancement of 1 further confirmed the structure of 1. In the 1H NMR spectrum, the signal of H-13 displayed a broad singlet, thus the 13-hydroxy group was determined to be α.10 The other relative stereochemistry of 1 was assigned as occurring with that of tenvermectins A and B.7 So, the structure of 1 was assigned as 13α-hydroxy-4-ethy1 milbemycin β3.
Compounds 2 and 3 were also obtained as colorless oil. The 1H and 13C NMR data (Table 1) of 1 and 2 were very similar to those of 1, which suggested that the three compounds possessed the same skeleton. The HMBC correlations from H3-32 to C-25, from H3-26 to C-3, C-4, C-5 and the cross peak of H3-32/H2-31 in the 1H-1H COSY spectrum (Figure 2) revealed that 2 is 13-hydroxy-25-ethy1 milbemycin β3. For 3, the HMBC correlations between H3-26 and C-3, C-4, C-5, between H-31 and C-24, C-25 and the 1H-1H COSY correlation of H3-31/H-25 (Figure 2) established the structure of 3 to be 13-hydroxy milbemycin β3. The relative configurations of 2 and 3 were assigned by analogy to 1.
Biological activity
The three new β-class milbemycins 1–3 possess potent acaricidal activity (Table 2). Especially, 1 was more active than 2, 3 and milbemycins A3/A4 mixture against larval mites. This result further demonstrated C-26 substituted milbemycins possessing high activity.11
Materials and methods
General
UV spectra were obtained on a Varian CARY 300 BIO spectrophotometer (Varian, Palo Alto, CA, USA); IR spectra were recorded on a Nicolet Magna FT-IR 750 spectrometer (Nicolet Magna, Madison, WI, USA); 1H and 13C NMR spectra were measured with a Bruker DRX-400 (400 MHz for 1H and 100 MHz for 13C) spectrometer (Bruker, Rheinstetten, Germany). Chemical shifts are reported in p.p.m. (δ), using residual CHCl3 (δH 7.26 ppm; δC 77.0) as an internal standard, with coupling constants (J) in Hz. 1H and 13C NMR assignments were supported by 1H-1H COSY, HMQC and HMBC experiments. The ESI-MS and high resolution electrospray ionization (HRESI-MS) spectra were taken on a Q-TOF Micro LC-MS-MS mass spectrometer (Waters, Milford, MA, USA). Optical rotation was measured on a Perkin-Elmer 341 polarimeter (Perkin-Elmer, Fremont, CA, USA). Commercial silica gel (Qing Dao Hai Yang Chemical Group Co., 100–200 mesh). Spots were detected on TLC under UV or by heating after spraying with sulfuric acid-ethanol, 5:95 (v/v).
Microorganism
Avermectin is produced by S. avermitilis and milbemycin can be produced by Streptomyces hygroscopicus. Both avermectin and milbemycin are biosynthesized by polyketide synthases. The C22–25 substructures of avermectin and milbemycin are determined by aveA1 and milA1, respectively. So plasmid used for replacing aveA1 gene with milA1 gene was constructed and it was transformed into S avermitilis G8–17, an avermectin B industrial strain. After subculture of a transformant, the genetically engineered strain MHJ1011, whose aveA1 gene was replaced by milA1 gene seamlessly, was screened.7
Fermentation
The strain S. avermitilis MHJ1011 was maintained on the medium containing glucose 10 g, malt extract (Bei Jing Ao Bo Xing, Beijing, China) 3 g, yeast extract (Bei Jing Ao Bo Xing) 3 g, K2HPO4·3H2O 0.5 g, MgSO4·7H2O 0.5 g, NaCl 0.5 g, KNO3 1 g and agar 20 g in 1.0 l of tap water, pH 7.0. The seed medium consisted of corn starch (Shanghai Guoqiang Bioengineering Equipment, Shanghai, China) 25.0 g, soybean meal (Ningbo Beilun Jiangnan Grease Co., Ltd, Ningbo, China) 8.0 g, peanut meal (Bei Jing Ao Bo Xing) 10.0 g, yeast extract 9.5 g, CoCl2·6H2O 0.03 g in 1-liter water and pH 7.4. All the media were sterilized at 121 °C for 20 min. Slant culture was incubated for 6–7 days at 28 °C. Fermentation was carried out in 50 l of fermentor (containing 30 l of production medium). The producing medium was composed of corn starch 14%, soybean meal 0.2%,α-amylase (Bei Jing Ao Bo Xing) 0.003%, yeast extract 1.0%, zeolite powder (Kaixi, Jiande City, Zhejiang, China) 0.2%, CoCl2·6H2O 0.002%, MnSO4 0.0024%, NaMoO4 0.0024% and pH 7.4. The fermentation was conducted at 28 °C for 10 days stirred at 100 r.p.m with an aeration rate of 900 l of air per hour.
Isolation and purification
The final 30 l of fermentation broth was filtered and the resulting cake was extracted with ethanol (10 l). The ethanol extract was evaporated under reduced pressure to 1 l at 45 °C and subsequently extracted three times using an equal volume of ethyl acetate. The combined ethyl acetate phase was concentrated under reduced pressure to yield 260 g of oily substances. A amount of 5 g of the residual oily substance was subjected to a silica gel column and successively eluted with a stepwise gradient of petroleum ether/acetone (90:10–60:40, v/v) to afford four fractions (I–IV) based on the (TLC profiles. The fraction I eluted with petroleum ether/acetone (90:10–80:20, v/v) was chromatographed on silica gel and eluted with petroleum ether-acetone (100:0–70:30, v/v) to give eight fractions. The fourth fraction was separated by the semi-preparative HPLC (Agilent 1100, Zorbax SB-C18, 5 μm, 250 × 9.4 mm inner diameter; Agilent, Palo Alto, CA, USA) using a solvent containing a CH3CN: H2O mixture (95: 5, v/v) to obtain compounds 1 (tR 15.1 min, 80 mg), 2 (tR 14.2 min, 40 mg) and 3 (tR 13.5 min, 35 mg).
13α-hydroxy-4-ethy1 milbemycin β3 (1): colorless oil;  (c 0.13, EtOH); UV (EtOH) λmax nm (log ɛ): 247 (4.17); IR (KBr), νmax cm−1: 3396, 1703, 2929, 1382, 1281, 1162, 994; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Table 1; ESI-MS m/z 525 [M+H]+; HRESI-MS m/z 525.3206 [M+H]+ (calcd for C32H45O6 525.3211).
(c 0.13, EtOH); UV (EtOH) λmax nm (log ɛ): 247 (4.17); IR (KBr), νmax cm−1: 3396, 1703, 2929, 1382, 1281, 1162, 994; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Table 1; ESI-MS m/z 525 [M+H]+; HRESI-MS m/z 525.3206 [M+H]+ (calcd for C32H45O6 525.3211).
13α-hydroxy-25-ethy1 milbemycin β3 (2): colorless oil;  (c 0.07, EtOH); UV (EtOH) λmax nm (log ɛ): 247 (4.33); IR (KBr), νmax cm−1: 3395, 1714, 2928, 1455, 1163, 996; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Table 1; ESI-MS m/z 525 [M+H]+; HRESI-MS m/z 525.3205 [M+H]+ (calcd for C32H45O6 525.3211).
(c 0.07, EtOH); UV (EtOH) λmax nm (log ɛ): 247 (4.33); IR (KBr), νmax cm−1: 3395, 1714, 2928, 1455, 1163, 996; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Table 1; ESI-MS m/z 525 [M+H]+; HRESI-MS m/z 525.3205 [M+H]+ (calcd for C32H45O6 525.3211).
13α-hydroxy milbemycin β3 (3): colorless oil,  (c 0.17, EtOH); UV (EtOH) λmax nm (log ɛ): 245 (3.97); IR (KBr), νmax cm−1: 3409, 1702, 2928, 1382, 1279, 1163, 997; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Table 1; ESI-MS m/z 509 [M-H]−; HRESI-MS m/z 509.2908 [M-H]− (calcd for C31H41O6 509.2909).
(c 0.17, EtOH); UV (EtOH) λmax nm (log ɛ): 245 (3.97); IR (KBr), νmax cm−1: 3409, 1702, 2928, 1382, 1279, 1163, 997; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data see Table 1; ESI-MS m/z 509 [M-H]−; HRESI-MS m/z 509.2908 [M-H]− (calcd for C31H41O6 509.2909).
Acaricidal activity test
The larvicidal activities of test compounds against Tetranychus cinnabarinus reared in the laboratory were tested according to the reported procedure.12 Each test sample was prepared in acetone at a concentration of 1000 mg l−1 and diluted to the required concentration of 0.01, 0.005, 0.0025, 0.001 and 0.0005 mg l−1 with distilled water containing alkylphenol ethoxylates (1/1000). The primary leaves of Vicia faba L. species were infected with carmine spider mites. At 2 h after infection, 10 fourth-instar mite larvae were dipped in the diluted solutions of related chemicals for 5 s before the superfluous liquid was removed and larvae were kept in a conditioned room. The experiments were repeated three times and blank controls. The mortality was evaluated 24 h after treatment by examining the adult mites under a binocular microscope to determine the living and dead individuals.
References
Wong, F. T. & Khosla, C. Combinatorial biosynthesis of polyketides-a perspective. Curr. Opin. Chem. Biol. 16, 117–123 (2012).
McDaniel, R. et al. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel ‘unnatural’ natural products. Proc. Natl Acad. Sci. USA 96, 1846–1851 (1999).
Rowe, C. J. et al. Engineering a polyketide with a longer chain by insertion of an extra module into the erythromycin-producing polyketide synthase. Chem. Biol. 8, 475–485 (2001).
Carmody, M. et al. Analysis and manipulation of amphotericin biosynthetic genes by means of modified phage KC515 transduction techniques. Gene. 343, 107–115 (2004).
Khan, N., Rawlings, B. & Caffrey, P. A labile point in mutant amphotericin polyketide synthases. Biotechnol. Lett. 33, 1121–1126 (2011).
Kopp, F. & Marahiel, M. A. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat. Prod. Rep. 24, 735–749 (2007).
Huang, J. et al. Gene replacement for the generation of designed novel avermectin derivatives with enhanced acaricidal and nematicidal activities. Appl. Environ. Microbiol. 81, 5326–5334 (2015).
Takiguchi, Y. et al. Milbemycins, a new family of macrolide antibiotics: fermentation, isolation and physic-chemical properties. J. Antibiot. 33, 1120–1127 (1980).
Xiang, W. S., Wang, J. D., Wang, X. J. & Zhang, J. Two new milbemycins from Streptomyces bingchenggensis: fermentation, isolation, structure elucidation and biological properties. J. Antibiot. 60, 351–356 (2007).
Blizzard, T. A. et al. Synthesis and biological activity of 13-epi-avermectins: potent anthelmintic agents with an increased margin of safety. J. Med. Chem. 35, 3873–3878 (1992).
Takahashi, S., Miyaoka, H., Tanaka, K., Enokita, R. & Okazaki, T. Milbemycins α11α12α13α14, and α15: a new family of milbemycins from Streptomyces hygroscopicus subsp. aureolacrimosus. J. Antibiot. 46, 1364–1371 (1993).
Sertkaya, E., Kaya, K. & Soylu, S. Acaricidal activities of the essential oils from several medicinal plants against the carmine spider mite. Ind. Crop. Prod. 31, 107–112 (2010).
Acknowledgements
This research work was financially supported by the National Natural Science Foundation of China (31471809).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Pan, Jj., Wan, X., Zhang, H. et al. Three new milbemycins from a genetically engineered strain S. avermitilis MHJ1011. J Antibiot 69, 104–107 (2016). https://doi.org/10.1038/ja.2015.90
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2015.90