Abstract
Proteus mirabilis rods are one of the most commonly isolated species of the Proteus genus from human infections, mainly those from the urinary tract and wounds. They are often related to biofilm structure formation. The bacterial cells of the biofilm are less susceptible to routinely used antimicrobials, making the treatment more difficult. The aim of this study was to evaluate quantitatively the influence of ceftazidime and ciprofloxacin on biofilm formation on the polyvinyl chloride surface by 42 P. mirabilis strains isolated from urine, purulence, wound swab and bedsore samples. It has been shown that ceftazidime and ciprofloxacin at concentrations equal to 1/4, 1/2 and 1 times their MIC values for particular Proteus spp. strains decrease their ability to form biofilms. Moreover, ciprofloxacin at concentrations equal to 1/4, 1/2 and 1 times their MIC values for particular P. mirabilis strains reduces biofilm formation more efficiently than ceftazidime at the corresponding concentration values.
Similar content being viewed by others
Introduction
Proteus spp. rods belong to the opportunistic human pathogens. The most frequently isolated species are P. mirabilis, and less common ones P. vulgaris, P. penneri and P. hauseri representatives. Proteus spp. rods significantly contribute to urinary, respiratory and digestive tract infections and skin infections, and, though less commonly, are isolated from bloodstream infections.1, 2 These rods can be characterized by the presence of different virulence factors, including biofilm structure formation.3, 4, 5
Biofilm is defined as a complex community of surface-associated cells enclosed in a polymer matrix containing open water channels.6 It can be described as a sessile community with cells that are irreversibly attached to a substratum or interface (or to each other), are embedded in a matrix of extracellular polymeric substances, water and noncellular or abiotic components. A definition for biofilm must therefore take into consideration the physiological attributes of the microorganisms, including altered phenotype and gene transcription.6
The process of biofilm formation can be divided into the following steps:6, 7, 8 initial attachment to a surface—nonspecific connection, specific adhesion—microcolony formation, growing up—development of a three-dimensional community structure, maturation and migration—detachment. A mature biofilm has a typical structure, consisting of three layers. The internal one is located directly on the basal material surface (inert or living) and contains tightly aggregated bacterial cells. The surface layer is placed directly on it and both of them are isolated from the environment by the external layer, characterized by the presence of rapidly metabolizing cells.
Biofilms are considered to be highly resistant to antimicrobial agents. The limited activity of most antibiotics and disinfectants against the microorganisms on the external biofilm surface7 is one of the causes for the resistance of the whole biofilm to antimicrobials.9, 10, 11, 12 Another cause is the metabolism type of the cells present in the structure of the biofilm.12 Bacterial cells living in the deeper biofilm layers have limited access to nutritional elements. This results in slower metabolism rate and overall bacterial growth. Antibiotics, whose half-time and doses are usually selective for planktonic cells and their metabolism type, do not have an effective concentration that is long enough to cause therapeutic success—eradication of the whole biofilm structure-embedded microorganisms pool.12 Antibiotic resistance in a biofilm could also be due to other mechanisms: permeability barrier, activation of resistance genes and development of resistance forms. These mechanisms may exist in a bacterial biofilm simultaneously.12
In these situations it is crucial to estimate the subinhibitory antimicrobials’ concentration (subminimal inhibitory concentration, subMIC) and their effect on biofilm cells because in vivo growing microorganisms are usually treated with such antibiotic concentrations.12 The basic element of bacterial biofilm resistance is the generation and selection of the persister cells—a small percentage of the bacteria that are resistant to administered antibiotics and capable of microbial population regeneration after unsuccessful antimicrobial treatment.11
The aim of this study was to evaluate the effects of ciprofloxacin’s and ceftazidime’s subMIC values on P. mirabilis rods’ ability to form biofilms on the surface of polyvinyl chloride, commonly used in urological catheter production.
Materials and methods
Material
A total number of 42 ciprofloxacin- and ceftazidime-susceptible P. mirabilis strains were included in the study. The examined strains were isolated from urine (10; 23.8%), wound swabs (19; 45.2%), bedsore swabs (7; 16.7%) and purulence (6; 14.3%) samples derived from the patients hospitalized between 2005 and 2009 in Dr Antoni Jurasz University Hospital No. 1 in Bydgoszcz—20 (47.6%) and 22 (52.4%) from women and men, respectively (Figure 1). Only two (4.8%) of the examined strains were involved in catheter-related infections. These strains were isolated from the urological catheters’ surfaces.
Susceptibility
The extended spectrum beta-lactamase (ESBL) synthesis tests were performed according to EUCAST recommendations (http://www.eucast.org/antimicrobial_susceptibility_testing/).
The ceftazidime and ciprofloxacin MIC values for each strain were determined by the Etest (AB BioDisk, Solna, Sweden) according to the manufacturer’s instructions and serial antibiotic dilution in liquid medium. Results obtained by applying both methods were comparable. Both antibiotics’ subMIC values were then subsequently chosen for checking their effects on P. mirabilis biofilm formation.
Genetic similarity
The examined Proteus spp. strains’ genetic similarity was evaluated by pulsed field gel electrophoresis using SfiI enzyme (Fermentas, Glen Burnie, MD, USA) restriction cleavage, as previously described by Sabbuba et al.13 in our own modifications.
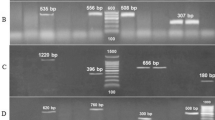
Briefly, the examined strains were cultured on MacConkey Agar (Becton Dickinson, Franklin Lakes, NJ, USA) for 24 h at 37 °C. One colony of each strain was harvested and suspended in 3 ml of tryptic soy broth medium (Bio-Rad, Hercules, CA, USA). After 24-hour incubation at 37 °C, each culture was centrifuged at 10 000 r.p.m. for 10 min. The supernatant was discarded and the pellet was resuspended in 100 μl of 0.5 M EDTA. The suspension was then incubated for 15 min at room temperature and centrifuged once again using the same parameters. The supernatant was discharged and the pellet was resuspended in 100 μl of SE buffer (0.1 M Tris-HCl/0.1 M EDTA, Sigma-Aldrich, St Louis, MO, USA). Twenty microliters of proteinase K (DNA, Gdańsk, Poland) and 170 μl of 2.0% low melting point (Bio-Rad) agarose solution with 1.0% of SDS (Sigma-Aldrich) at a temperature of 45 °C were added subsequently and 20 μl discs were formed. After 30 min at 4 °C the discs were solidified and subsequently treated with 1 ml of lytic buffer (0.1 M EDTA, Bio-Rad; 1.0% N-lauryl-sarcosyl, Sigma-Aldrich; 0.1 M Tris-HCl, Sigma-Aldrich) with the addition of 12 μl of proteinase K. After a 2-h digestion at 54 °C the discs were washed twice with sterile deionized water at a temperature of 55 °C and three times with 1 ml of Tris-EDTA buffer at 50 °C (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The discs were then put for 30 min into 100 μl of Tango buffer (Fermentas) and a 3-h restriction cleavage in a volume of 50 μl of G buffer (Fermentas) with 10 U of SfiI enzyme was done. The discs, containing digested DNA, were loaded into the wells of 1.0% agarose gel prepared in 0.5 × tris-boric acid-EDTA (Bio-Rad), and a molecular size marker (100–970 kbp) was added into the border wells and sealed with agarose. Bacterial DNA fragments were then separated at 12 °C for 24 h at 6 V cm−1 with a switch time of 1–30 s using CHEF-DR II (Bio-Rad) apparatus. After separation, the gels were stained with ethidium bromide, washed, observed in UV light and photographed (Figure 1). Visualizations of the stained gels were collected on Gel Doc 2000 program Quantity One 4.1.1. (Bio-Rad) and analyzed with Molecular Analyst Fingerprinting (Bio-Rad). The dendrograms showing the relatedness of all of the examined P. mirabilis strains were constructed afterwards.
Biofilm measurement
P. mirabilis rods’ biofilm formation ability was evaluated in vitro on the polyvinyl chloride surface of the urological catheters (Unomedical, Birkerød, Denmark). Sterile biomaterial fragments (1 cm) were obtained by cutting the catheters in aseptic conditions. For the contamination control of this step, a biomaterial fragment was placed into tryptic soy broth medium for 24-hour incubation at 37 °C. P. mirabilis biofilm formation was assessed using Richards’ method, described by Różalska et al.14 The obtained results were interpreted statistically. All the values below the average value minus 0.5 of standard deviation were interpreted as weak biofilm formation, values between the average and average minus 0.5 standard deviationas moderate, values between average and average plus 0.5 standard deviationas strong, and those above upper average plus 0.5 standard deviation as very strong biofilm formation.
To evaluate the impact of the chosen antibiotics on biofilm formation, 24-hour P. mirabilis cultures at 37 °C on MacConkey Agar were done. Suspensions of each bacterial strain, adjusted to a density of 1.0 in McFarland’s scale in a volume of 10 ml of tryptic soy broth each, were done from the obtained cultures. Two milliliters of each suspension was placed separately into five sterile tubes and the biomaterial fragment was added subsequently and incubated for 24 h at 37 °C. The catheters covered with 24-h biofilm were washed twice in 5 ml of the sterile phosphate-buffered saline (pH 7.2, BTL, Łódź, Poland) solution and moved to sterile Mueller-Hinton broth (Becton Dickinson) containing ceftazidime (Sigma-Aldrich) or ciprofloxacin (Sigma-Aldrich) added according to the 1, 1/2, 1/4 and 1/8 MIC values of the antibiotics for the particular strain.
The biomaterial fragments covered with the biofilm of each strain were placed into antibiotic-free Mueller-Hinton broth and served as control. ATCC 29213 Staphylococcus aureus strain was chosen for evaluation of ceftazidime and ciprofloxacin MIC values. A sterile biomaterial fragment placed into the Mueller-Hinton broth served as sterility control.
The examined strains were incubated for 24 h at 37 °C. The biofilm-covered biomaterial fragments of each strain were then washed 3–5 times with sterile phosphate-buffered saline, placed into 1 ml of 0.5% saponin solution (Fluka Analytical, Busch, Switzerland) and shaken at 1100 r.p.m. for 60 min at 37 °C. The obtained bacterial suspensions in saponin were used to prepare a bacterial dilution (10 times each) series and seeded quantitatively onto three tryptic soy agar (Bio-Rad) plates for each strain. Results were read out after 24-hour incubation at 37 °C and expressed as colony-forming units per milliliter (CFU ml−1). The medium value was calculated from three cultures of each dilution. Results between 30 and 300 colonies per plate and plates without single colonies growth type were excluded. The rate of biofilm killing achieved by antibiotics usage was calculated according to the formula:
Biofilm killing percentage , where Aw is the CFU ml−1 value obtained from antibiotic-free medium, and Aa the CFU ml−1 value obtained from antibiotic-containing medium.
, where Aw is the CFU ml−1 value obtained from antibiotic-free medium, and Aa the CFU ml−1 value obtained from antibiotic-containing medium.
Statistical analysis
The obtained results’ normality was evaluated using Shapiro–Wilk test. Statistical analysis on the antibiotics’ effect on biofilm formation was done with non-parametric Friedman and Kruskal–Wallis analysis of variance test and with post-hoc test (Bonferoni). The differences in biofilm formation between ESBL(+) and ESBL(−) strains were analyzed using Mann–Whitney U-test. Differences were considered to be statistically significant when α⩽0.05.
Results
Susceptibility
Thirteen (30.9%) of the studied strains were ESBL-positive. The strains involved in the catheter-related infection were ESBL-negative.
Genetic similarity
The examined strains’ pulsed field gel electrophoresis patterns analysis did not reveal existence of the genetically indistinguishable strains (Figure 1). Four pairs of the strains included into the study presented a value ⩾80% of the genetic similarity. The strains presented as number 29 and 86, with the highest rate of genetic relatedness, also had similar biofilm formation rates and the same antibiotics susceptibility pattern but different ESBL-synthesis ability. The rest of the examined strains gave distinct profiles with genetic relatedness in the range of 40–80%.
Biofilm formation
According to the established criteria for biofilm formation ability, of 42 examined strains, 16 (38.1%) strains formed biofilm weakly, 10 (23.8%) moderately, 4 (9.5%) strongly and 12 (28.6%) very strongly.
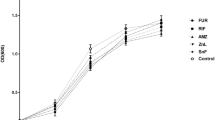
Addition of both ceftazidime and ciprofloxacin to the proliferation medium for 24 h at concentrations equal to 1/4, 1/2 and 1 times the MIC values inhibited biofilm formation by P. mirabilis (Figure 2).
The biofilm killing rate for ceftazidime at concentrations equal to 1/8, 1/4, 1/2 and 1 times the MIC values were 22.9%, 64.4%, 83.3% and 95.3%, respectively.
Ciprofloxacin addition reduced biofilm formation at concentrations equal to 1/4, 1/2 and 1 times the MIC values and the corresponding biofilm killing rates were 88.6%, 98.9% and 99.8%, respectively. The chosen fluoroquinolone at a concentration of 1/8 MIC caused ∼22.4% increase in biofilm formation.
The differences obtained for the effect of ceftazidime and ciprofloxacin addition on biofilm formation at 1/4, 1/2 and 1 times their MIC values, when compared with the antibiotic-free medium, were statistically significant (P values of Wilcoxon’s test were 0.0009, 0.0000, 0.0000 and 0.0001, 0.0000, 0.0000, respectively). We did not find any influence of 1/8 ceftazidime and ciprofloxacin MIC values on biofilm formation (P values reached 0.1972 and 0.3409, respectively). Besides, we found that the growing concentration of the applied antibiotics is statistically more effective in biofilm killing (P value <0.001 in each case).
ESBL(+) strains formed biofilms slightly weakly in antibiotic-free medium when compared with their ESBL(−) counterparts and the observed differences were not statistically significant (Table 1). CFU ml−1 median values for ESBL(+) P. mirabilis strains forming a biofilm on polyvinyl chloride surface in the presence of ceftazidime at each applied concentration were higher when compared with their ESBL(−) counterparts (Table 1).
In the current study the differences in ceftazidime’s and ciprofloxacin’s effect on different P. mirabilis strains’ biofilm synthesis intensity were also evaluated (Figure 3a and b). Among moderately biofilm-forming strains in the medium containing ceftazidime at a concentration equal to 1/8 of its MIC values, higher CFU ml−1 median values (125.5 × 106) were obtained when compared with antibiotic-free medium of the corresponding value (107.0 × 106). Similar accuracy but stronger expression was observed for strongly and very strongly biofilm-forming strains cultured in the presence of 1/8 ciprofloxacin MIC (256.0 × 106 and 321.0 × 106 CFU ml−1, respectively). The median values of the results obtained for the strains cultured in either subMIC or MIC values for both antibiotics were lower than those obtained in the antibiotic-free medium cultures.
The effects of ciprofloxacin on biofilm killing in the examined strains at a concentration equal to 1/8 its MIC values, depending on its overall synthesis intensity, revealed statistically significant difference (P=0.0138). Very strongly biofilm-forming strains reacted in a weaker manner than moderately intensive biofilm structure-forming strains when ciprofloxacin at a concentration equal to its 1/8 MIC value was added.
At the applied ceftazidime subMIC values, none of the differences obtained for the biofilm-forming killing were statistically significant, in terms of strains with different biofilm-forming intensity.
Discussion
Treatment of the clinical outcomes related to biofilm formation is a crucial problem of modern medicine. The biofilm cells display increased resistance to antimicrobials when compared with planktonic ones. During the antibiotic treatment, the biofilm-embedded cells are treated mainly at subliminal antimicrobial concentrations, which may change the bacterial cells’ structure and properties. For example, usage of ceftazidime, azithromycin and ciprofloxacin at subMIC values leads to the loss of E. coli’s ability to adhere to intermediate epithelium.15
Ceftazidime at a concentration equal to 1/16 of its MIC value leads to a situation where 60% of the E. coli cells can no longer bind to epithelial cells. Increase in subMIC value correlates with the decreased number of bacterial cells binding to intermediate epithelium. The decreased cell number of three E. coli strains adhering to human vaginal epithelial cells was also observed after ampicillin, ceftazidime, gentamicin, ciprofloxacin and co-trimoxazole usage at the subMIC values.16 Wojnicz and Jankowski17 proved that in an environment with 1/8, 1/4, and 1/2 of the ciprofloxacin MIC values, the number of E. coli cells adhering to the epithelial cell surface decreases by 50.0–70.0% when compared with chemotherapeutic-free environmental conditions. Moreover, E. coli cells treated with ciprofloxacin MIC values are more susceptible to bactericidal complement activity.18 This is most probably related to the outer membrane protein structure changes.
Bret and Di Martino19 have not shown any significant differences in the chosen antibiotic’s effect on biofilm-forming strains, regardless of their mode of action and subMIC values for particular strains. In the presence of ceftazidime and ciprofloxacin at concentrations equal to their subMIC values, both E. coli and P. vulgaris strains formed biofilms less intensively on the polystyrene surface when compared with the antibiotic-free growth conditions in the control group. In the present study, depending on the choice of antibiotic (different in their antimicrobial mode of action) differences in Proteus spp. strains’ biofilm-forming abilities were observed. Ceftazidime and ciprofloxacin addition at concentrations equal to 1 and 1/2 of their MIC values for the particular strain led to an increase in the bacterial cell number isolated from the biofilm structure. Usage of 1 and 1/2 times ciprofloxacin’s MIC values decreased Proteus spp. strains’ biofilm formation on the polyvinyl chloride more effectively when compared with ceftazidime. Similar results were obtained by Wasfi et al.20 on evaluating the different antibiotics’ effect on four P. mirabilis strains’ biofilm-forming abilities. Among the four antimicrobials included in the study (ceftriaxone, ciprofloxacin, nitrofurantoin and gentamicin), the highest biofilm formation-limiting properties were observed for ciprofloxacin. It suggests a better biofilm-forming limitation effect for this representative of the antimicrobial groups that disrupt DNA synthesis than for those afflicted with bacterial cell wall synthesis. It might have also resulted from fluoroquinolones’ capacity to penetrate the extracellular polymeric substance structure.21
The limitation of E. coli cells’ adhesion to epithelial cells on addition of ceftazidime, azithromycin and ciprofloxacin at their subMIC values is related to bacterial cell wall changes, including dissection along layers and surface layer depletion.15 Moreover, fluoroquinolones decrease the extracellular matrix synthesis.22 P. mirabilis rods’ biofilm-forming limitation may also be related to decreased flagella formation. This was confirmed by the study of Horii et al.23 They indicated that mupirocin at subMIC values decreases expression of the P. mirabilis’ virulence factor genes, including those encoded for cilia synthesis.
The antibiotics may also influence the virulence factor-coding genes’ expression through the quorum sensing phenomenon. Skindersoe et al.24 revealed that ceftazidime, azithromycin and ciprofloxacin reduce P. aeruginosa virulence factor-coding genes’ expression using N-acyl-L-homoserine lactones. Moreover, Jones et al.25 proved the quorum sensing inhibitors’ effect on biofilm formation using P. mirabilis strain. Both tannic acid and p-nitrophenyl glycerol were observed to reduce biofilm-forming cells’ number in artificial urine by three logarithmic units and, simultaneously, decrease urine pH value from 8.3–8.4 to 6.1–6.7, respectively.
To summarize, ceftazidime and ciprofloxacin at concentrations corresponding to 1, 1/2 and 1/4 times their MIC values limit P. mirabilis rods’ biofilm formation on the surface of polyvinyl chloride. Moreover, ciprofloxacin at concentrations equal to 1/4, 1/2 and 1 times its MIC value presents stronger biofilm-forming limitation abilities when compared with corresponding ceftazidime concentration values, whereas at low concentration (1/8 MIC) it may stimulate biofilm formation by P. mirabilis strains.
References
Endimiani, A. et al. Proteus mirabilis bloodstream infections: risk factors and treatment outcome related to the expression of extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 49, 2598–2605 (2005).
O’Hara, C. M., Brenner, F. W. & Miller, J. M. Classification, identification and clinical significance of Proteus, Providencia and Morganella. Clin. Microbiol. Rev. 13, 534–546 (2000).
Champs, C., Bonnet, R., Sirot, D., Chanal, C. & Sirot, J. Clinical relevance of Proteus mirabilis in hospital patients: a two year survey. J. Antimicrob. Chemother. 45, 537–539 (2000).
Coker, C. h., Poore, C. A., Li, X. & Mobley, H. L. T. Pathogenesis of Proteus mirabilis urinary tract infection. Microb. Infect. 2, 1497–1505 (2000).
Różalski, A., Kwil, I., Torzewska, A., Baranowska, M. & Strączek, P. Proteus bacilli: features and virulence factors. Post. Hig. Med. Dośw. 61, 204–219 (2007).
Donlan, R. M. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 15, 1387–1392 (2001).
Karatan, E. & Watnick, P. Signals, regulatory networks and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347 (2009).
Prakash, B., Veeregowda, B. M. & Krishnappa, G. Biofilms: a survival strategy of bacteria. Curr. Sci. 85, 1299–1306 (2003).
Burmoølle, M. et al. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 72, 3916–3923 (2006).
Chambless, J. D., Hunt, S. M. & Stewart, P. S. A three-dimensional computer model of four hypothetical mechanisms protecting biofilms from antimicrobials. Appl. Environ. Microbiol. 72, 2005–2013 (2006).
Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45, 999–1007 (2001).
Stewart, P. S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292, 107–113 (2002).
Sabbuba, N. A., Mahenthiralingam, E. & Stickler, D. J. Molecular epidemiology of Proteus mirabilis infections of the catheterized urinary tract. J. Clin. Microbiol. 41, 4961–4965 (2003).
Różalska, B., Sadowska., B., Więckowska, M. & Rudnicka, W. Detection of bacterial biofilm on medical biomaterials. Med. Dośw. Mikrobiol. 50, 115–122 (1998).
Vranes, J. Effect of subinhibitory concentrations of ceftazidime. Ciprofloxacin and azithromycin on the hemoagglutination and adherence of uropathoenic Escherichia coli strains. Chemotherapy 42, 177–185 (1996).
Vidya, K. C., Mallya, P. S. & Pao, P. S. Inhibition of bacterial adhesion by subinhibitory concentration of antibiotics. Ind. J. Med. Microbiol. 23, 102–105 (2005).
Wojnicz, D. & Jankowski, S. Effects of subinhibitory concentrations of amikacin and ciprofloxacin on the hydrophobicity and adherence to epithelial cells of uropathogenic Escherichia coli strains. Int. J. Antimicrob. Agents 29, 700–704 (2007).
Wojnicz, D. & Cisowska, A. Composition of outer membrane proteins of Escherichia coli strains in relation to serum susceptibility after exposure to subinhibitiry concentrations of amikacin and ciprofloxacin. Int. J. Antimicrob. Agents 33, 579–582 (2009).
Bret, L. & Di Martino, P. Effect of ceftazidime, amikacin and ciprofloxacin on biofilm formation by some enterobacterial clinical isolates. Chemotherapy 50, 255–259 (2004).
Wasfi, R. et al. Antimicrobial activities against biofilm formed by Proteus mirabilis isolates from wound and urinary tract infections. Ind. J. Med. Microbiol. 30, 76–80 (2012).
Wang, D., Wang, Y. & Liu, Y. Activity of ciprofloxacin and azithromycin on biofilms produced in vitro by Haemophilus influenza. Chin. Med. J. 122, 1305–1310 (2009).
Di Bonaventura, G., Specicato, I., D’Antonio, D., Robuffo, I. & Piccolomini, R. Biofilm formation by Stenotrophomonas maltophilia: modulation by quinolones, trimethoprim-sulfamethoxazole and ceftazidime. Antimicrob. Agents Chemother. 48, 151–160 (2004).
Horii, T. et al. Effects of mupirocin at subinhibitory concentrations on flagella formation in Pseudomonas aeruginosa and Proteus mirabilis. J. Antimicrob. Chemother. 51, 1175–1179 (2003).
Skindersoe, M. E. et al. Effect of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52, 3648–3663 (2008).
Jones, S. M., Dang, T. T. & Martinuzzi, R. Use of quorum sensing antagonists to deter the formation of crystalline Proteus mirabilis biofilms. Int. J. Antimicrob. Agents 34, 360–364 (2009).
Acknowledgements
The study was funded by Nicolaus Copernicus University grant number 09/2008 and The Integrated Regional Development Program 2009.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kwiecińska-Piróg, J., Bogiel, T. & Gospodarek, E. Effects of ceftazidime and ciprofloxacin on biofilm formation in Proteus mirabilis rods. J Antibiot 66, 593–597 (2013). https://doi.org/10.1038/ja.2013.59
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.59